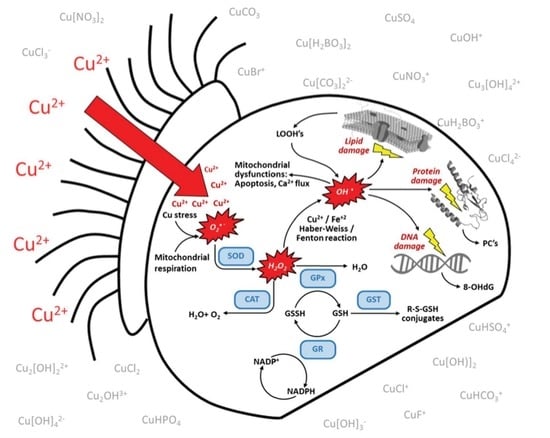
Metabolic Regulation of Copper Toxicity during Marine Mussel Embryogenesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design Summary
2.2. Chemicals and Seawater Preparation
2.3. Broodstock Collection and Spawning
2.4. Fertilisation and Tank Incubation
2.5. Embryo and Larval Sampling
2.6. Seawater Chemistry
2.6.1. Bulk Seawater Composition
2.6.2. Copper Speciation Analysis
2.7. Metabolite Analysis
2.7.1. Sample Preparation
2.7.2. GC-MS Analysis
2.7.3. Data Pre-Processing and Metabolite Identification
2.7.4. Statistical Analysis and Data Visualisation
2.8. Oxidative Stress/Damage Analysis
2.8.1. Protein, Lipid, and DNA Analysis
2.8.2. Antioxidant Enzyme Analysis
2.8.3. Reduced Glutathione Analysis
2.8.4. Analysis of Reactive Oxygen Species
2.8.5. Statistics and Data Presentation
3. Results
3.1. Seawater Chemistry
3.2. Survival and Development
3.3. Untargeted Metabolomics Analyses
3.3.1. Lethal Exposure Effects
3.3.2. Sublethal Exposure Effects
3.4. Targeted Analysis of the ROS Regulatory System
4. Discussion
4.1. Seawater Chemistry
4.2. Biochemical Profiling
4.2.1. Energy Metabolism
4.2.2. Amino Acid Metabolism
4.2.3. Osmoregulation
4.2.4. Neurotoxicity
4.2.5. Pyrimidine Catabolism
4.2.6. Oxidative Stress, Redox Homeostasis, and Metal Chelation
4.2.7. Lipid Metabolism
4.2.8. Immunotoxic Associations
4.2.9. Other Metabolic Responses
4.3. Metabolomics as a Health Assessment Tool
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Uriu-Adams, J.Y.; Keen, C.L. Copper, oxidative stress, and human health. Mol. Asp. Med. 2005, 26, 268–298. [Google Scholar]
- Lee, J.; Prohaska, J.R.; Thiele, D.J. Essential role for mammalian copper transporter Ctr1 in copper homeostasis and embryonic development. Proc. Natl. Acad. Sci. USA 2001, 98, 6842–6847. [Google Scholar]
- Madsen, E.; Gitlin, J.D. Copper deficiency. Curr. Opin. Gastroenterol. 2007, 23, 187–192. [Google Scholar] [PubMed]
- Horn, D.; Barrientos, A. Mitochondrial copper metabolism and delivery to cytochrome c oxidase. IUBMB Life 2008, 60, 421–429. [Google Scholar] [PubMed] [Green Version]
- Nose, Y.; Rees, E.M.; Thiele, D.J. Structure of the Ctr1 copper trans ‘PORE’ter reveals novel architecture. Trends Biochem. Sci. 2006, 31, 604–607. [Google Scholar]
- Klotz, L.O.; Kröncke, K.D.; Buchczyk, D.P.; Sies, H. Role of copper, zinc, selenium and tellurium in the cellular defense against oxidative and nitrosative stress. J. Nutr. 2003, 133, 1448S–1451S. [Google Scholar]
- Scheiber, I.F.; Mercer, J.F.; Dringen, R. Metabolism and functions of copper in brain. Prog. Neurobiol. 2014, 116, 33–57. [Google Scholar]
- Harrison, M.D.; Jones, C.E.; Solioz, M.; Dameron, C.T. Intracellular copper routing: The role of copper chaperones. Trends Biochem. Sci. 2000, 25, 29–32. [Google Scholar] [CrossRef]
- Dennery, P.A. Effects of oxidative stress on embryonic development. Birth Defects Res. Part C Embryo Today Rev. 2007, 81, 155–162. [Google Scholar]
- Temple, M.D.; Perrone, G.G.; Dawes, I.W. Complex cellular responses to reactive oxygen species. Trends Cell Biol. 2005, 15, 319–326. [Google Scholar]
- Puig, S.; Thiele, D.J. Molecular mechanisms of copper uptake and distribution. Curr. Opin. Chem. Biol. 2002, 6, 171–180. [Google Scholar]
- Kim, B.E.; Nevitt, T.; Thiele, D.J. Mechanisms for copper acquisition, distribution and regulation. Nat. Chem. Biol. 2008, 4, 176–185. [Google Scholar]
- Rubino, J.T.; Franz, K.J. Coordination chemistry of copper proteins: How nature handles a toxic cargo for essential function. J. Inorg. Biochem. 2012, 107, 129–143. [Google Scholar] [PubMed]
- Schöne, B.R.; Krause, R.A. Retrospective environmental biomonitoring–Mussel Watch expanded. Glob. Planet. Chang. 2016, 144, 228–251. [Google Scholar]
- Farrington, J.W.; Tripp, B.W.; Tanabe, S.; Subramanian, A.; Sericano, J.L.; Wade, T.L.; Knap, A.H.; Edward, D. Goldberg’s proposal of “the mussel watch”: Reflections after 40 years. Mar. Pollut. Bull. 2016, 110, 501–510. [Google Scholar] [PubMed]
- Arnold, W.; Cotsifas, J.S.; Ogle, R.S.; DePalma, S.G.; Smith, D.S. A comparison of the copper sensitivity of six invertebrate species in ambient salt water of varying dissolved organic matter concentrations. Environ. Toxicol. Chem. 2010, 29, 311–319. [Google Scholar]
- Funes, V.; Alhama, J.; Navas, J.I.; López-Barea, J.; Peinado, J. Ecotoxicological effects of metal pollution in two mollusc species from the Spanish South Atlantic littoral. Environ. Pollut. 2006, 139, 214–223. [Google Scholar]
- Neff, J.M. Copper in seawater. In Bioaccumulation in Marine Organisms: Effect of Contaminants from Oil Well Produced Water; Neff, J.M., Ed.; Elsevier: Amsterdam, The Netherlands, 2002; pp. 145–160. [Google Scholar]
- Gadd, J.; Cameron, M. Antifouling Biocides in Marinas: Measurement of Copper Concentrations and Comparison to Model Predictions for Eight Auckland Sites; Auckland Council Technical Report TR2012/033; NIWA for Auckland Council: Auckland, New Zealand, 2012. [Google Scholar]
- Jahan, S.; Strezov, V. Water quality assessment of Australian ports using water quality evaluation indices. PLoS ONE 2017, 12, e0189284. [Google Scholar]
- ANZECC; ARMCANZ. Australian Guidelines for Water Quality Monitoring and Reporting; National Water Quality Management Strategy Paper No 7; Australian and New Zealand Environment and Conservation Council (ANZECC) & Agriculture and Resource Management Council of Australia and New Zealand (ARMCANZ): Canberra, Australia, 2000.
- Martin, M.; Osborn, K.E.; Billig, P.; Glickstein, N. Toxicities of ten metals to Crassostrea gigas and Mytilus edulis embryos and Cancer magister larvae. Mar. Pollut. Bull. 1981, 12, 305–308. [Google Scholar]
- Rosen, G.; Rivera-Duarte, I.; Kear-Padilla, L.; Chadwick, D.B. Use of laboratory toxicity tests with bivalve and echinoderm embryos to evaluate the bioavailability of copper in San Diego Bay, California, USA. Environ. Toxicol. Chem. 2005, 24, 415–422. [Google Scholar] [CrossRef] [Green Version]
- Nadella, S.R.; Fitzpatrick, J.L.; Franklin, N.; Bucking, C.; Smith, S.; Wood, C.M. Toxicity of dissolved Cu, Zn, Ni and Cd to developing embryos of the blue mussel (Mytilus trossolus) and the protective effect of dissolved organic carbon. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2009, 149, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Bosse, C.; Rosen, G.; Colvin, M.; Earley, P.; Santore, R.; Rivera-Duarte, I. Copper bioavailability and toxicity to Mytilus galloprovincialis in Shelter Island Yacht Basin, San Diego, CA. Mar. Pollut. Bull. 2014, 85, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Worboys, M.A.; Leung, K.M.Y.; Grist, E.P.M.; Crane, M. Time should be considered in developmental ecotoxicity test. Mar. Pollut. Bull. 2002, 45, 92–99. [Google Scholar] [CrossRef] [PubMed]
- King, C.K.; Riddle, M.J. Effects of metal contaminants on the development of the common Antarctic sea urchin Sterechinus neumayeri and comparisons of sensitivity with tropical and temperate echinoids. Mar. Ecol. Prog. Ser. 2001, 215, 143–154. [Google Scholar] [CrossRef]
- Beca Infrastructure. Auckland Recreational Boating Study; Beca Infrastructure Ltd. (Beca) for Auckland Council: Auckland, New Zealand, 2012. [Google Scholar]
- Gadd, J.; Depree, C.; Hickey, C. Relevance to New Zealand of the OECD Emission Scenario Document for Antifouling Paints: Phase 2 Report; Report for the Environmental Protection Authority (EPA); National Institute of Water and Atmospheric Research Ltd.: Hamilton, New Zealand, 2011. [Google Scholar]
- Smith, P.J. Selective decline in imposex levels in the dogwhelk Lepsiella scobina following a ban on the use of TBT antifoulants in New Zealand. Mar. Pollut. Bull. 1996, 32, 362–365. [Google Scholar] [CrossRef]
- Symonds, J.E.; Clarke, S.M.; King, N.; Walker, S.P.; Blanchard, B.; Sutherland, D.; Roberts, R.; Preece, M.A.; Tate, M.; Buxton, P.; et al. Developing successful breeding programs for New Zealand aquaculture: A perspective on progress and future genomic opportunities. Front. Genet. 2019, 10, 27. [Google Scholar] [CrossRef] [Green Version]
- Bradley, N. SPATnz End of Programme Review Primary Growth Partnership: SPATnz Programme Evaluation Report; Report prepared for Ministry for Primary Industries; Maven Consulting Ltd.: North Vancouver, BC, Canada, 2020. [Google Scholar]
- Chandurvelan, R.; Marsden, I.D.; Gaw, S.; Glover, C.N. Impairment of green-lipped mussel (Perna canaliculus) physiology by waterborne cadmium: Relationship to tissue bioaccumulation and effect of exposure duration. Aquat. Toxicol. 2012, 124, 114–124. [Google Scholar] [CrossRef]
- Deruytter, D.; Vandegehuchte, M.B.; Garrevoet, J.; De Laender, F.; Vergucht, E.; Delbeke, K.; Blust, R.; De Schamphelaere, K.A.; Vincze, L.; Janssen, C.R. Salinity and dissolved organic carbon both affect copper toxicity in mussel larvae: Copper speciation or competition cannot explain everything. Environ. Toxicol. Chem. 2015, 34, 1330–1336. [Google Scholar] [CrossRef]
- McDougall, D.R.; Chan, A.; McGillivray, D.J.; de Jonge, M.D.; Miskelly, G.M.; Jeffs, A.G. Examining the role of ethylenediaminetetraacetic acid (EDTA) in larval shellfish production in seawater contaminated with heavy metals. Aquat. Toxicol. 2019, 217, 105330. [Google Scholar] [CrossRef]
- Gale, S.L.; Burritt, D.J.; Adams, S.L. The role of ethylenediaminetetraacetic acid in green-lipped mussel (Perna canaliculus) embryo development: A biochemical and morphological characterization. Aquaculture 2016, 463, 22–27. [Google Scholar] [CrossRef]
- McDougall, D.R.; Kihara, S.; Reinhardt, J.; Miskelly, G.M.; McGillivray, D.J.; Jeffs, A.G. Biodegradable chelating agent improves the survival of early larvae for shellfish aquaculture. Aquat. Toxicol. 2020, 228, 105645. [Google Scholar] [CrossRef] [PubMed]
- Ragg, N.L.; Gale, S.L.; Le, D.V.; Hawes, N.A.; Burritt, D.J.; Young, T.; Ericson, J.A.; Hilton, Z.; Watts, E.; Berry, J.; et al. The Effects of aragonite saturation state on hatchery-reared larvae of the Greenshell mussel Perna canaliculus. J. Shellfish Res. 2019, 38, 779–793. [Google Scholar] [CrossRef]
- Rusk, A.B.; Alfaro, A.C.; Young, T.; Watts, E.; Adams, S.L. Investigation of early mussel (Perna canaliculus) development using histology, SEM imaging, immunochemistry and confocal microscopy. Mar. Biol. Res. 2017, 13, 314–329. [Google Scholar] [CrossRef]
- Young, T.; Alfaro, A.C.; Sánchez-Lazo, C.; Robertson, J. Putative involvement of adrenergic receptors in regulation of mussel (Perna canaliculus) larval settlement. Mar. Biol. Res. 2015, 11, 655–665. [Google Scholar] [CrossRef] [Green Version]
- Howard, D.W.; Smith, C.S. Histological Techniques for Marine Bivalve Mollusks; NOAA Technical Memorandum NMFS-F/NEC-25; National Oceanic and Atmospheric Administration: Woods Hole, MA, USA, 1983.
- Zitoun, R.; Clearwater, S.J.; Hassler, C.; Thompson, K.J.; Albert, A.; Sander, S.G. Copper toxicity to blue mussel embryos (Mytilus galloprovincialis): The effect of natural dissolved organic matter on copper toxicity in estuarine waters. Sci. Total Environ. 2019, 653, 300–314. [Google Scholar] [CrossRef]
- Stewart, M.; Cameron, M.; McMurtry, M.; Sander, S.G.; Benedict, B.; Graham, L.; Hosie, M.; Green, T. Development of passive sampling devices for bioavailable contaminants of current and emerging concern: Waitemata Harbour case study. N. Z. J. Mar. Freshw. Res. 2016, 50, 526–548. [Google Scholar] [CrossRef]
- Gustafsson, J.P. Visual MINTEQ Ver. 3.0; Based on de Allison JD, Brown DS, Novo-Gradac KJ, MINTEQA2 ver, 4.; KTH Department of Land and Water Resources Engineering: Stockholm, Sweden, 2011. [Google Scholar]
- Sander, S.G.; Buck, K.N.; Wells, M. The effect of natural organic ligands on trace metal speciation in San Francisco Bay: Implications for water quality criteria. Mar. Chem. 2015, 173, 269–281. [Google Scholar] [CrossRef]
- Villas-Bôas, S.G.; Smart, K.F.; Sivakumaran, S.; Lane, G.A. Alkylation or silylation for analysis of amino and non-amino organic acids by GC-MS? Metabolites 2011, 1, 3–20. [Google Scholar] [CrossRef] [Green Version]
- Smart, K.F.; Aggio, R.B.; Van Houtte, J.R.; Villas-Bôas, S.G. Analytical platform for metabolome analysis of microbial cells using MCF derivatization followed by gas chromatography-mass spectrometry. Nat. Protoc. 2010, 5, 1709–1729. [Google Scholar] [CrossRef]
- Aggio, R.; Villas-Bôas, S.G.; Ruggiero, K. Metab: An R package for high-throughput analysis of metabolomics data generated by GC-MS. Bioinformatics 2011, 27, 2316–2318. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.; Mandal, R.; Sinelnikov, I.V.; Broadhurst, D.; Wishart, D.S. MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, T.; Kesarcodi-Watson, A.; Alfaro, A.C.; Merien, F.; Nguyen, T.V.; Mae, H.; Le, D.V.; Villas-Bôas, S. Differential expression of novel metabolic and immunological biomarkers in oysters challenged with a virulent strain of OsHV-1. Dev. Comp. Immunol. 2017, 73, 229–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, R.; Wang, J.; Su, M.; Jia, E.; Chen, S.; Chen, T.; Ni, Y. Missing value imputation approach for mass spectrometry-based metabolomics data. Sci. Rep. 2018, 8, 663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caraux, G.; Pinloche, S. PermutMatrix: A graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics 2005, 21, 1280–1281. [Google Scholar] [CrossRef] [Green Version]
- Fryer, H.J.; Davis, G.E.; Manthorpe, M.; Varon, S. Lowry protein assay using an automatic microtiter plate spectrophotometer. Anal. Biochem. 1986, 153, 262–266. [Google Scholar] [CrossRef]
- Reznick, A.Z.; Packer, L. Oxidative damage to proteins: Spectrophotometric method for carbonyl assay. Methods Enzymol. 1994, 233, 357–363. [Google Scholar]
- Mihaljević, B.; Katušin-Ražem, B.; Ražem, D. The reevaluation of the ferric thiocyanate assay for lipid hydroperoxides with special considerations of the mechanistic aspects of the response. Free Radic. Biol. Med. 1996, 21, 53–63. [Google Scholar] [CrossRef]
- Banowetz, G.M.; Dierksen, K.P.; Azevedo, M.D.; Stout, R. Microplate quantification of plant leaf superoxide dismutases. Anal. Biochem. 2004, 332, 314–320. [Google Scholar] [CrossRef]
- Janssens, B.J.; Childress, J.J.; Baguet, F.; Rees, J.F. Reduced enzymatic antioxidative defense in deep-sea fish. J. Exp. Biol. 2000, 203, 3717–3725. [Google Scholar] [CrossRef]
- Paglia, D.E.; Valentine, W.N. Studies on the quantitative and qualitative characterization of erythrocyte glutathione peroxidase. J. Lab. Clin. Med. 1967, 70, 158–169. [Google Scholar]
- Cribb, A.E.; Leeder, J.S.; Spielberg, S.P. Use of a microplate reader in an assay of glutathione reductase using 5,5′-dithiobis (2-nitrobenzoic acid). Anal. Biochem. 1989, 183, 195–196. [Google Scholar] [CrossRef] [PubMed]
- Habig, W.H.; Pabst, M.J.; Jakoby, W.B. Glutathione S-transferases the first enzymatic step in mercapturic acid formation. J. Biol. Chem. 1974, 249, 7130–7139. [Google Scholar] [CrossRef] [PubMed]
- Kogut, M.B.; Voelker, B.M. Strong copper-binding behavior of terrestrial humic substances in seawater. Environ. Sci. Technol. 2001, 35, 1149–1156. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, J.I.; Beiras, R.; Mubiana, V.K.; Blust, R. Copper uptake by Mytilus edulis in the presence of humic acids. Environ. Toxicol. Chem. 2005, 24, 973–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helm, M.M.; Bourne, N.; Lovatelli, A. Hatchery Culture of Bivalves: A Practical Manual; FOA Fisheries Technical Paper; Food and Agriculture Organization of the United Nations: Rome, Italy, 2004. [Google Scholar]
- Hasegawa, H.; Lind, E.J.; Boin, M.A.; Häse, C.C. The extracellular metalloprotease of Vibrio tubiashii is a major virulence factor for pacific oyster (Crassostrea gigas) larvae. Appl. Environ. Microbiol. 2008, 74, 4101–4110. [Google Scholar] [CrossRef] [Green Version]
- Hasegawa, H.; Häse, C.C. The extracellular metalloprotease of Vibrio tubiashii directly inhibits its extracellular haemolysin. Microbiology 2009, 155, 2296–2305. [Google Scholar] [CrossRef]
- Mersni-Achour, R.; Cheikh, Y.B.; Pichereau, V.; Doghri, I.; Etien, C.; Degremont, L.; Saulnier, D.; Fruitier-Arnaudin, I.; Travers, M.-A. Factors other than metalloprotease are required for full virulence of French Vibrio tubiashii isolates in oyster larvae. Microbiology 2015, 161, 997–1007. [Google Scholar] [CrossRef]
- Wang, W.; Lv, X.; Liu, Z.; Song, X.; Yi, Q.; Wang, L.; Song, L. The sensing pattern and antitoxic response of Crassostrea gigas against extracellular products of Vibrio splendidus. Dev. Comp. Immunol. 2020, 102, 103467. [Google Scholar] [CrossRef]
- Sweetlove, L.J.; Beard, K.F.; Nunes-Nesi, A.; Fernie, A.R.; Ratcliffe, R.G. Not just a circle: Flux modes in the plant TCA cycle. Trends Plant Sci. 2010, 15, 462–470. [Google Scholar] [CrossRef]
- Cheng, T.; Sudderth, J.; Yang, C.; Mullen, A.R.; Jin, E.S.; Matés, J.M.; DeBerardinis, R.J. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc. Natl. Acad. Sci. USA 2011, 108, 8674–8679. [Google Scholar] [CrossRef]
- Chan, E.C.Y.; Koh, P.K.; Mal, M.; Cheah, P.Y.; Eu, K.W.; Backshall, A.; Cavill, R.; Nicholson, J.K.; Keun, H.C. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS). J. Proteome Res. 2009, 8, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Aggio, R.B.; Ruggiero, K.; Villas-Bôas, S.G. Pathway Activity Profiling (PAPi): From the metabolite profile to the metabolic pathway activity. Bioinformatics 2010, 26, 2969–2976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morita, M.; Gravel, S.P.; Chénard, V.; Sikström, K.; Zheng, L.; Alain, T.; Gandin, V.; Avizonis, D.; Arguello, M.; Zakaria, C.; et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 2013, 18, 698–711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romanowska, E.; Igamberdiev, A.U.; Parys, E.; Gardeström, P. Stimulation of respiration by Pb2+ in detached leaves and mitochondria of C3 and C4 plants. Physiol. Plant. 2002, 116, 148–154. [Google Scholar] [CrossRef]
- Vosloo, A.; van Aardt, W.J.; Mienie, L.J. Sublethal effects of copper on the freshwater crab Potamonautes warreni. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2002, 133, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Handy, R.D. Chronic effects of copper exposure versus endocrine toxicity: Two sides of the same toxicological process? Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2003, 135, 25–38. [Google Scholar] [CrossRef]
- Muyssen, B.T.; de Schamphelaere, K.A.; Janssen, C.R. Mechanisms of chronic waterborne Zn toxicity in Daphnia magna. Aquat. Toxicol. 2006, 77, 393–401. [Google Scholar] [CrossRef]
- Cheng, Z.; Tsuda, M.; Kishita, Y.; Sato, Y.; Aigaki, T. Impaired energy metabolism in a Drosophila model of mitochondrial aconitase deficiency. Biochem. Biophys. Res. Commun. 2013, 433, 145–150. [Google Scholar] [CrossRef]
- Tretter, L.; Adam-Vizi, V. Inhibition of Krebs cycle enzymes by hydrogen peroxide: A key role of α-ketoglutarate dehydrogenase in limiting NADH production under oxidative stress. J. Neurosci. 2000, 20, 8972–8979. [Google Scholar] [CrossRef] [Green Version]
- Bota, D.A.; Davies, K.J. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat. Cell Biol. 2002, 4, 674–680. [Google Scholar] [CrossRef]
- Talbot, D.A.; Brand, M.D. Uncoupling protein 3 protects aconitase against inactivation in isolated skeletal muscle mitochondria. Biochim. Biophys. Acta Bioenerg. 2005, 1709, 150–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cherkasov, A.A.; Overton, R.A.; Sokolov, E.P.; Sokolova, I.M. Temperature-dependent effects of cadmium and purine nucleotides on mitochondrial aconitase from a marine ectotherm, Crassostrea virginica: A role of temperature in oxidative stress and allosteric enzyme regulation. J. Exp. Biol. 2007, 210, 46–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lushchak, O.V.; Piroddi, M.; Galli, F.; Lushchak, V.I. Aconitase post-translational modification as a key in linkage between Krebs cycle, iron homeostasis, redox signaling, and metabolism of reactive oxygen species. Redox Rep. 2014, 19, 8–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winterbourn, C.C. Toxicity of iron and hydrogen peroxide: The Fenton reaction. Toxicol. Lett. 1995, 82, 969–974. [Google Scholar] [CrossRef]
- Murakami, K.; Yoshino, M. Inactivation of aconitase in yeast exposed to oxidative stress. IUBMB Life 1997, 41, 481–486. [Google Scholar] [CrossRef]
- Kehrer, J.P. The Haber–Weiss reaction and mechanisms of toxicity. Toxicology 2000, 149, 43–50. [Google Scholar] [CrossRef]
- Cantu, D.; Schaack, J.; Patel, M. Oxidative inactivation of mitochondrial aconitase results in iron and H2O2-mediated neurotoxicity in rat primary mesencephalic cultures. PLoS ONE 2009, 4, e7095. [Google Scholar] [CrossRef] [Green Version]
- Rose, S.; Melnyk, S.; Pavliv, O.; Bai, S.; Nick, T.G.; Frye, R.E.; James, S.J. Evidence of oxidative damage and inflammation associated with low glutathione redox status in the autism brain. Transl. Psychiatry 2012, 2, e134. [Google Scholar] [CrossRef] [Green Version]
- Rønnestad, I.; Fyhn, H.J. Metabolic aspects of free amino acids in developing marine fish eggs and larvae. Rev. Fish. Sci. 1993, 1, 239–259. [Google Scholar] [CrossRef]
- Rønnestad, I.; Tonheim, S.K.; Fyhn, H.J.; Rojas-Garcı, C.R.; Kamisaka, Y.; Koven, W.; Finn, R.; Terjesen, B.; Barr, Y.; Conceição, L. The supply of amino acids during early feeding stages of marine fish larvae: A review of recent findings. Aquaculture 2003, 227, 147–164. [Google Scholar] [CrossRef]
- Da Costa, F.; Nóvoa, S.; Ojea, J.; Martínez-Patiño, D. Changes in biochemical and fatty acid composition of the razor clam Solen marginatus (Solenidae: Bivalvia) during larval development. Mar. Biol. 2011, 158, 1829–1840. [Google Scholar] [CrossRef]
- Waterlow, J. Protein Turnover; CABI Publishing: Wallingford, UK, 2006. [Google Scholar]
- Combs, G.F.; Trumbo, P.R.; McKinley, M.C.; Milner, J.; Studenski, S.; Kimura, T.; Watkins, S.M.; Raiten, D.J. Biomarkers in nutrition: New frontiers in research and application. Ann. N. Y. Acad. Sci. 2013, 1278, 1–10. [Google Scholar] [CrossRef]
- Wu, H.C.; Chen, H.M.; Shiau, C.Y. Free amino acids and peptides as related to antioxidant properties in protein hydrolysates of mackerel (Scomber austriasicus). Food Res. Int. 2003, 36, 949–957. [Google Scholar] [CrossRef]
- Elias, R.J.; Kellerby, S.S.; Decker, E.A. Antioxidant activity of proteins and peptides. Crit. Rev. Food Sci. Nutr. 2008, 48, 430–441. [Google Scholar] [CrossRef]
- Hyde, R.; Taylor, P.M.; Hundal, H.S. Amino acid transporters: Roles in amino acid sensing and signalling in animal cells. Biochem. J. 2003, 373, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meijer, A.J.; Dubbelhuis, P.F. Amino acid signalling and the integration of metabolism. Biochem. Biophys. Res. Commun. 2004, 313, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Kube, S.; Gerber, A.; Jansen, J.M.; Schiedek, D. Patterns of organic osmolytes in two marine bivalves, Macoma balthica, and Mytilus spp.; along their European distribution. Mar. Biol. 2006, 149, 1387–1396. [Google Scholar] [CrossRef]
- Powell, E.N.; Kasschau, M.; Chen, E.; Koenig, M.; Pecon, J. Changes in the free amino acid pool during environmental stress in the gill tissue of the oyster, Crassostrea virginica. Comp. Biochem. Physiol. Part A Physiol. 1982, 71, 591–598. [Google Scholar] [CrossRef]
- Sansone, G.; Cotugno, M.; Cosma, I.; Zatta, P. The effect of β-alanine on the concentration of taurine and other free amino acids during osmotic stress of Mytilus galloprovincialis. J. Mar. Biol. Assoc. U. K. 1987, 67, 111–117. [Google Scholar] [CrossRef]
- Hosoi, M.; Kubota, S.; Toyohara, M.; Toyohara, H.; Hayashi, I. Effect of salinity change on free amino acid content in Pacific oyster. Fish. Sci. 2003, 69, 395–400. [Google Scholar] [CrossRef]
- Preston, R.L. Transport of amino acids by marine invertebrates. J. Exp. Zool. 1993, 265, 410–421. [Google Scholar] [CrossRef]
- Grosell, M.; Blanchard, J.; Brix, K.V.; Gerdes, R. Physiology is pivotal for interactions between salinity and acute copper toxicity to fish and invertebrates. Aquat. Toxicol. 2007, 84, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wang, W.X. NMR-based metabolomic studies on the toxicological effects of cadmium and copper on green mussels Perna viridis. Aquat. Toxicol. 2010, 100, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, X.; You, L.; Zhou, D.; Wu, H.; Li, L.; Zhao, J.; Feng, J.; Yu, J. Metabolic responses in gills of Manila clam Ruditapes philippinarum exposed to copper using NMR-based metabolomics. Mar. Environ. Res. 2011, 72, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.K.; Jung, Y.S.; Park, J.C.; Seo, J.; Choi, M.S.; Hwang, G.S. Characterizing the effect of heavy metal contamination on marine mussels using metabolomics. Mar. Pollut. Bull. 2012, 64, 1874–1879. [Google Scholar] [CrossRef]
- Ji, C.; Wang, Q.; Wu, H.; Tan, Q.; Wang, W.-X. A metabolomic investigation of the effects of metal pollution in oysters Crassostrea hongkongensis. Mar. Pollut. Bull. 2015, 90, 317–322. [Google Scholar] [CrossRef]
- Ji, C.; Cao, L.; Li, F. Toxicological evaluation of two pedigrees of clam Ruditapes philippinarum as bioindicators of heavy metal contaminants using metabolomics. Environ. Toxicol. Pharmacol. 2015, 39, 545–554. [Google Scholar] [CrossRef]
- Lopes, T.M.; Barcarolli, I.F.; de Oliveira, C.B.; de Souza, M.M.; Bianchini, A. Effect of copper on ion content in isolated mantle cells of the marine clam Mesodesma mactroides. Environ. Toxicol. Chem. 2011, 30, 1582–1585. [Google Scholar] [CrossRef]
- Nogueira, L.S.; Wood, C.M.; Gillis, P.L.; Bianchini, A. Isolation and fractionation of gill cells from freshwater (Lasmigona costata) and seawater (Mesodesma mactroides) bivalves for use in toxicological studies with copper. Cytotechnology 2013, 65, 773–783. [Google Scholar] [CrossRef] [Green Version]
- Boyle, R.T.; Oliveira, L.F.; Bianchini, A.; Souza, M.M. The effects of copper on Na+/K+-ATPase and aquaporin expression in two euryhaline invertebrates. Bull. Environ. Contam. Toxicol. 2013, 90, 387–390. [Google Scholar] [CrossRef]
- Jorge, M.B.; Lauer, M.M.; Martins, C.D.M.G.; Bianchini, A. Impaired regulation of divalent cations with acute copper exposure in the marine clam Mesodesma mactroides. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2016, 179, 79–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bishop, S.H.; Greenwalt, D.E.; Kapper, M.A.; Paynter, K.T.; Ellis, L.L. Metabolic regulation of proline, glycine, and alanine accumulation as intracellular osmolytes in ribbed mussel gill tissue. J. Exp. Zool. 1994, 268, 151–161. [Google Scholar] [CrossRef]
- Pierce, S.K. Osmolyte permeability in molluscan red cells is regulated by Ca2+ and membrane protein phosphorylation: The present perspective. J. Exp. Zool. 1994, 268, 166–170. [Google Scholar] [CrossRef]
- Alfaro, A.C.; Young, T.; Ganesan, A.M. Regulatory effects of mussel (Aulacomya maoriana Iredale 1915) larval settlement by neuroactive compounds, amino acids and bacterial biofilms. Aquaculture 2011, 322, 158–168. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Lazo, C.; Martínez-Pita, I.; Young, T.; Alfaro, A.C. Induction of settlement in larvae of the mussel Mytilus galloprovincialis using neuroactive compounds. Aquaculture 2012, 344–349, 210–215. [Google Scholar] [CrossRef] [Green Version]
- Alfaro, A.C.; Young, T.; Bowden, K. Neurophysiological control of swimming behaviour, attachment and metamorphosis in black-footed abalone (Haliotis iris) larvae. N. Z. J. Mar. Freshw. Res. 2014, 48, 314–334. [Google Scholar] [CrossRef] [Green Version]
- Pires, A.; Croll, R.P.; Hadfield, M.G. Catecholamines modulate metamorphosis in the opisthobranch gastropod Phestilla sibogae. Biol. Bull. 2000, 198, 319–331. [Google Scholar] [CrossRef]
- Boadle-Biber, M.C.; Roth, R.H. Factors modifying the synthesis of dopamine from tyrosine in pedal ganglia of Mercenaria mercenaria (Mollusca). Comp. Gen. Pharmacol. 1972, 3, 61–74. [Google Scholar] [CrossRef]
- Zhu, W.; Mantione, K.J.; Shen, L.; Cadet, P.; Esch, T.; Goumon, Y.; Bianchi, E.; Sonetti, D.; Stefano, G.B. Tyrosine and tyramine increase endogenous ganglionic morphine and dopamine levels in vitro and in vivo: Cyp2d6 and tyrosine hydroxylase modulation demonstrates a dopamine coupling. Med. Sci. Monit. 2005, 11, BR397–BR404. [Google Scholar]
- Di Poi, C.; Evariste, L.; Serpentini, A.; Halm-Lemeille, M.P.; Lebel, J.M.; Costil, K. Toxicity of five antidepressant drugs on embryo–larval development and metamorphosis success in the Pacific oyster, Crassostrea gigas. Environ. Sci. Pollut. Res. 2014, 21, 13302–13314. [Google Scholar] [CrossRef]
- Sudama, G.; Zhang, J.; Isbister, J.; Willett, J.D. Metabolic profiling in Caenorhabditis elegans provides an unbiased approach to investigations of dosage dependent lead toxicity. Metabolomics 2013, 9, 189–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poynton, H.C.; Taylor, N.S.; Hicks, J.; Colson, K.; Chan, S.; Clark, C.; Scanlan, L.; Loguinov, A.V.; Vulpe, C.; Viant, M.R. Metabolomics of microliter hemolymph samples enables an improved understanding of the combined metabolic and transcriptional responses of Daphnia magna to cadmium. Environ. Sci. Technol. 2011, 45, 3710–3717. [Google Scholar] [CrossRef] [PubMed]
- Nagato, E.G.; Jessica, C.; Lankadurai, B.P.; Poirier, D.G.; Reiner, E.J.; Simpson, A.J.; Simpson, M.J. 1H NMR-based metabolomics investigation of Daphnia magna responses to sub-lethal exposure to arsenic, copper and lithium. Chemosphere 2013, 93, 331–337. [Google Scholar] [CrossRef]
- Dickinson, A.J.; Croll, R.P.; Voronezhskaya, E.E. Development of embryonic cells containing serotonin, catecholamines, and FMRFamide-related peptides in Aplysia californica. Biol. Bull. 2000, 199, 305–315. [Google Scholar] [CrossRef]
- Buznikov, G.A.; Nikitina, L.A.; Voronezhskaya, E.E.; Bezuglov, V.V.; Willows, A.D.; Nezlin, L.P. Localization of serotonin and its possible role in early embryos of Tritonia diomedea (Mollusca: Nudibranchia). Cell Tissue Res. 2003, 311, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Franchini, A. The distribution of cells containing FMRFamide-and 5-HT-related molecules in the embryonic development of Viviparus ater (Mollusca, Gastropoda). Eur. J. Histochem. 2005, 49, 301–308. [Google Scholar] [CrossRef]
- Filla, A.; Hiripi, L.; Elekes, K. Role of aminergic (serotonin and dopamine) systems in the embryogenesis and different embryonic behaviors of the pond snail, Lymnaea stagnalis. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2009, 149, 73–82. [Google Scholar] [CrossRef]
- Guerrier, P.; Leclerc-David, C.; Moreau, M. Evidence for the involvement of internal calcium stores during serotonin-induced meiosis reinitiation in oocytes of the bivalve mollusc Ruditapes philippinarum. Dev. Biol. 1993, 159, 474–484. [Google Scholar] [CrossRef]
- Kempf, S.C.; Page, L.R.; Pires, A. Development of serotonin-like immunoreactivity in the embryos and larvae of nudibranch mollusks with emphasis on the structure and possible function of the apical sensory organ. J. Comp. Neurol. 1997, 386, 507–528. [Google Scholar] [CrossRef]
- De Boeck, G.; Nilsson, G.E.; Elofsson, U.; Vlaeminck, A.; Blust, R. Brain monoamine levels and energy status in common carp (Cyprinus carpio) after exposure to sublethal levels of copper. Aquat. Toxicol. 1995, 33, 265–277. [Google Scholar] [CrossRef]
- Lepage, O.; Tottmar, O.; Winberg, S. Elevated dietary intake of L-tryptophan counteracts the stress-induced elevation of plasma cortisol in rainbow trout (Oncorhynchus mykiss). J. Exp. Biol. 2002, 205, 3679–3687. [Google Scholar] [CrossRef] [PubMed]
- Morandini, L.; Ramallo, M.R.; Moreira, R.G.; Höcht, C.; Somoza, G.M.; Silva, A.; Pandolfi, M. Serotonergic outcome, stress and sexual steroid hormones, and growth in a South American cichlid fish fed with an L-tryptophan enriched diet. Gen. Comp. Endocrinol. 2015, 223, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Hoseini, S.M.; Hosseini, S.A.; Soudagar, M. Dietary tryptophan changes serum stress markers, enzyme activity, and ions concentration of wild common carp Cyprinus carpio exposed to ambient copper. Fish Physiol. Biochem. 2012, 38, 1419–1426. [Google Scholar] [CrossRef] [PubMed]
- Fatahi, S.; Hoseini, S.M. Effect of dietary tryptophan and betaine on tolerance of Caspian roach (Rutilus rutilus caspicus) to copper toxicity. Int. J. Aquat. Biol. 2013, 1, 76–81. [Google Scholar]
- Mathieu, S.; Sylvain, D.; Walden, F.; Catapane, E.; Carroll, M. GABA is an inhibitory neurotransmitter in ganglia of the bivalve mollusc, Crassostrea virginica. FASEB J. 2014, 28 (Suppl. S1), 1059.4. [Google Scholar] [CrossRef]
- Stewart, P.; Soonklang, N.; Stewart, M.J.; Wanichanon, C.; Hanna, P.J.; Poomtong, T.; Sobhon, P. Larval settlement of the tropical abalone, Haliotis asinina Linnaeus, using natural and artificial chemical inducers. Aquac. Res. 2008, 39, 1181–1189. [Google Scholar] [CrossRef]
- Young, T.; Alfaro, A.C.; Robertson, J. Effect of neuroactive compounds on the settlement of mussel (Perna canaliculus) larvae. Aquaculture 2011, 319, 277–283. [Google Scholar] [CrossRef] [Green Version]
- Naidenko, T.K. Induction of metamorphosis of two species of sea urchin from Sea of Japan. Mar. Biol. 1996, 126, 685–692. [Google Scholar] [CrossRef]
- Sato, Y.; Yazaki, I. A cellular analysis of sea urchin metamorphosis induced by L-glutamine. In Echinoderm Research 1998; Carnevali, M.D.C., Bonasoro, F., Eds.; A.A. Balkema: Rotterdam, The Netherlands, 1999; pp. 221–226. [Google Scholar]
- Hatakeyama, D.; Aonuma, H.; Ito, E.; Elekes, K. Localization of glutamate-like immunoreactive neurons in the central and peripheral nervous system of the adult and developing pond snail, Lymnaea stagnalis. Biol. Bull. 2007, 213, 172–186. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Wang, W.X. Tissue-specific toxicological effects of cadmium in green mussels (Perna viridis): Nuclear magnetic resonance-based metabolomics study. Environ. Toxicol. Chem. 2011, 30, 806–812. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, L.; You, L.; Cong, M.; Zhao, J.; Wu, H.; Li, C.; Liu, D.; Yu, J. Toxicological responses to acute mercury exposure for three species of Manila clam Ruditapes philippinarum by NMR-based metabolomics. Environ. Toxicol. Pharmacol. 2011, 31, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, L.; You, L.; Yu, J.; Zhao, J.; Li, L.; Wang, Q.; Li, F.; Li, C.; Liu, D.; et al. Differential toxicological effects induced by mercury in gills from three pedigrees of Manila clam Ruditapes philippinarum by NMR-based metabolomics. Ecotoxicology 2011, 20, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Yoon, D.; Lee, M.; Yoon, C.; Kim, S. Metabolic responses in zebrafish (Danio rerio) exposed to zinc and cadmium by nuclear magnetic resonance-based metabolomics. Chem. Ecol. 2016, 32, 136–148. [Google Scholar] [CrossRef]
- Casanova, F.M.; Honda, R.T.; Ferreira-Nozawa, M.S.; Aride, P.H.R.; Nozawa, S.R. Effects of copper and cadmium exposure on mRnA expression of catalase, Glutamine synthetase, cytochrome p450 and Heat shock protein 70 in Tambaqui fish (Colossoma Macropomum). Gene Expr. Genet. Genom. 2013, 6, 1–8. [Google Scholar]
- Venkata, C.M.; Radhakrishnaiah, K. A study on the toxicity of cadmium on certain aspects of protein metabolism of the freshwater mussel Lamellidens marginalis (Lamarck) and freshwater fish Labeo rohita (Hamilton). Int. J. Environ. Sci. 2013, 4, 15–27. [Google Scholar]
- Fitsanakis, V.A.; Aschner, M. The importance of glutamate, glycine, and γ-aminobutyric acid transport and regulation in manganese, mercury and lead neurotoxicity. Toxicol. Appl. Pharmacol. 2005, 204, 343–354. [Google Scholar] [CrossRef]
- Sidoryk-Wegrzynowicz, M.; Aschner, M. Manganese toxicity in the central nervous system: The glutamine/glutamate-γ-aminobutyric acid cycle. J. Intern. Med. 2013, 273, 466–477. [Google Scholar] [CrossRef] [Green Version]
- Wirbisky, S.E.; Weber, G.J.; Lee, J.W.; Cannon, J.R.; Freeman, J.L. Novel dose-dependent alterations in excitatory GABA during embryonic development associated with lead (Pb) neurotoxicity. Toxicol. Lett. 2014, 229, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Awapara, J.; Allen, K. Occurrence of β-aminoisobutyric acid in Mytilus edulis. Science 1959, 130, 1250. [Google Scholar] [CrossRef]
- Nielsen, H.R.; Nyholm, K.; Sjølin, K.E. Relationship between urinary β-aminoisobutyric acid and transfer RNA turnover in cancer patients. Cancer Res. 1974, 34, 3428–3432. [Google Scholar]
- Farkas, W.R.; Fischbein, A.; Solomon, S.; Buschman, F.; Borek, E.; Sharma, O.K. Elevated urinary excretion of β-aminoisobutyric acid and exposure to inorganic lead. Arch. Environ. Health Int. J. 1987, 42, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Tomokuni, K.; Ichiba, M.; Hirai, Y. Elevated urinary excretion of β-aminoisobutyric acid and δ-aminolevulinic acid (ALA) and the inhibition of ALA-synthase and ALA-dehydratase activities in both liver and kidney in mice exposed to lead. Toxicol. Lett. 1991, 59, 169–173. [Google Scholar] [PubMed]
- Tomokuni, K.; Ichiba, M.; Mori, K. Relation between urinary beta-aminoisobutyric acid excretion and concentration of lead in the blood of workers occupationally exposed to lead. Br. J. Ind. Med. 1992, 49, 365–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brinzer, R.A.; Henderson, L.; Marchiondo, A.A.; Woods, D.J.; Davies, S.A.; Dow, J.A. Metabolomic profiling of permethrin-treated Drosophila melanogaster identifies a role for tryptophan catabolism in insecticide survival. Insect Biochem. Mol. Biol. 2015, 67, 74–86. [Google Scholar] [CrossRef] [Green Version]
- Van Kuilenburg, A.B.; Meinsma, R.; Beke, E.; Assmann, B.; Ribes, A.; Lorente, I.; Busch, R.; Mayatepek, E.; Abeling, N.G.; van Cruchten, A.; et al. β-Ureidopropionase deficiency: An inborn error of pyrimidine degradation associated with neurological abnormalities. Hum. Mol. Genet. 2004, 13, 2793–2801. [Google Scholar] [CrossRef]
- Van Kuilenburg, A.B.P.; Stroomer, A.E.M.; Abeling, N.G.G.M.; van Gennip, A.H. A pivotal role for β-aminoisobutyric acid and oxidative stress in dihydropyrimidine dehydrogenase deficiency? Nucl. Nucl. Nucleic Acids 2006, 25, 1103–1106. [Google Scholar] [CrossRef]
- Glahn, F.; Schmidt-Heck, W.; Zellmer, S.; Guthke, R.; Wiese, J.; Golka, K.; Hergenröder, R.; Degen, G.H.; Lehmann, T.; Hermes, M.; et al. Cadmium, cobalt and lead cause stress response, cell cycle deregulation and increased steroid as well as xenobiotic metabolism in primary normal human bronchial epithelial cells which is coordinated by at least nine transcription factors. Arch. Toxicol. 2008, 82, 513–524. [Google Scholar] [CrossRef]
- Gemelli, T.; de Andrade, R.B.; Rojas, D.B.; Bonorino, N.F.; Mazzola, P.N.; Tortorelli, L.S.; Funchal, C.; Dutra Filho, C.S.; Wannmacher, C.M.D. Effects of β-alanine administration on selected parameters of oxidative stress and phosphoryltransfer network in cerebral cortex and cerebellum of rats. Mol. Cell. Biochem. 2013, 380, 161–170. [Google Scholar] [CrossRef]
- Bo, Y.; Jin, C.; Liu, Y.; Yu, W.; Kang, H. Metabolomic analysis on the toxicological effects of TiO2 nanoparticles in mouse fibroblast cells: From the perspective of perturbations in amino acid metabolism. Toxicol. Mech. Methods 2014, 24, 461–469. [Google Scholar] [CrossRef]
- Liu, X.; Li, Q.; Sheng, J.; Hu, B.; Zhu, Z.; Zhou, S.; Yin, J.; Gong, Q.; Wang, Y.; Zhang, C. Unique plasma metabolomic signature of osteonecrosis of the femoral head. J. Orthop. Res. 2015, 34, 1158–1167. [Google Scholar] [CrossRef] [Green Version]
- Miccheli, A.; Capuani, G.; Marini, F.; Tomassini, A.; Praticò, G.; Ceccarelli, S.; Gnani, D.; Baviera, G.; Alisi, A.; Putignani, L.; et al. Urinary 1H-NMR-based metabolic profiling of children with NAFLD undergoing VSL#3 treatment. Int. J. Obes. 2015, 39, 1118–1125. [Google Scholar]
- Valavanidis, A.; Vlahogianni, T.; Dassenakis, M.; Scoullos, M. Molecular biomarkers of oxidative stress in aquatic organisms in relation to toxic environmental pollutants. Ecotoxicol. Environ. Saf. 2006, 64, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Sevcikova, M.; Modra, H.; Slaninova, A.; Svobodova, Z. Metals as a cause of oxidative stress in fish: A review. Vet. Med. 2011, 56, 537–546. [Google Scholar] [CrossRef] [Green Version]
- Copley, S.D.; Frank, E.; Kirsch, W.M.; Koch, T.H. Detection and possible origins of aminomalonic acid in protein hydrolysates. Anal. Biochem. 1992, 201, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.T.; Fu, S.; Stocker, R.; Davies, M.J. Biochemistry and pathology of radical-mediated protein oxidation. Biochem. J. 1997, 324, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Kang, D.K.; Jeong, J.; Drake, S.K.; Wehr, N.B.; Rouault, T.A.; Levine, R.L. Iron regulatory protein 2 as iron sensor Iron-dependent oxidative modification of cysteine. J. Biol. Chem. 2003, 278, 14857–14864. [Google Scholar] [CrossRef] [Green Version]
- Ibarra, R.; Dazard, J.E.; Sandlers, Y.; Rehman, F.; Abbas, R.; Kombu, R.; Zhang, G.-F.; Brunengraber, H.; Sanabria, J. Metabolomic analysis of liver tissue from the VX2 rabbit model of secondary liver tumors. HPB Surg. 2014, 2014, 310372. [Google Scholar] [CrossRef] [Green Version]
- Vandenbrouck, T.; Jones, O.A.; Dom, N.; Griffin, J.L.; De Coen, W. Mixtures of similarly acting compounds in Daphnia magna: From gene to metabolite and beyond. Environ. Int. 2010, 36, 254–268. [Google Scholar] [CrossRef]
- Gibb, J.O.T.; Svendsen, C.; Weeks, J.M.; Nicholson, J.K. 1H NMR spectroscopic investigations of tissue metabolite biomarker response to Cu II exposure in terrestrial invertebrates: Identification of free histidine as a novel biomarker of exposure to copper in earthworms. Biomarkers 1997, 2, 295–302. [Google Scholar] [CrossRef]
- Krämer, U.; Cotter-Howells, J.D.; Charnock, J.M.; Baker, A.J.; Smith, J.A.C. Free histidine as a metal chelator in plants that accumulate nickel. Nature 1996, 379, 635–638. [Google Scholar] [CrossRef]
- Wycisk, K.; Kim, E.J.; Schroeder, J.I.; Krämer, U. Enhancing the first enzymatic step in the histidine biosynthesis pathway increases the free histidine pool and nickel tolerance in Arabidopsis thaliana. FEBS Lett. 2004, 578, 128–134. [Google Scholar] [CrossRef]
- Bundy, J.G.; Sidhu, J.K.; Rana, F.; Spurgeon, D.J.; Svendsen, C.; Wren, J.F.; Stürzenbaum, S.R.; Morgan, A.J.; Kille, P. ‘Systems toxicology’ approach identifies coordinated metabolic responses to copper in a terrestrial non-model invertebrate, the earthworm Lumbricus rubellus. BMC Biol. 2008, 6, 25. [Google Scholar] [CrossRef] [Green Version]
- Digilio, G.; Sforzini, S.; Cassino, C.; Robotti, E.; Oliveri, C.; Marengo, E.; Musso, D.; Osella, D.; Viarengo, A. Haemolymph from Mytilus galloprovincialis: Response to copper and temperature challenges studied by 1H-NMR metabonomics. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2016, 183, 61–71. [Google Scholar] [CrossRef]
- Abebe, A.T.; Devoid, S.J.; Sugumaran, M.; Etter, R.; Robinson, W.E. Identification and quantification of histidine-rich glycoprotein (HRG) in the blood plasma of six marine bivalves. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2007, 147, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Devoid, S.J.; Etter, R.; Sugumaran, M.; Wallace, G.T.; Robinson, W.E. Histidine-rich glycoprotein from the hemolymph of the marine mussel Mytilus edulis L. binds Class A, Class B, and borderline metals. Environ. Toxicol. Chem. 2007, 26, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, Y.; Kashiba, M.; Maruyama, K.; Hoshikawa, K.; Sasaki, R.; Saito, K.; Kimura, H.; Goda, N.; Suematsu, M. Cadmium exposure alters metabolomics of sulfur containing amino acids in rat testes. Antioxid. Redox Signal. 2005, 7, 781–787. [Google Scholar] [CrossRef]
- Hughes, S.L.; Bundy, J.G.; Want, E.J.; Kille, P.; Sturzenbaum, S.R. The metabolomic responses of Caenorhabditis elegans to cadmium are largely independent of metallothionein status but dominated by changes in cystathionine and phytochelatins. J. Proteome Res. 2009, 8, 3512–3519. [Google Scholar] [CrossRef] [PubMed]
- Jeppe, K.J.; Carew, M.E.; Long, S.M.; Lee, S.F.; Pettigrove, V.; Hoffmann, A.A. Genes involved in cysteine metabolism of Chironomus tepperi are regulated differently by copper and by cadmium. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2014, 162, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Long, S.M.; Tull, D.L.; Jeppe, K.J.; De Souza, D.P.; Dayalan, S.; Pettigrove, V.J.; McConville, M.J.; Hoffmann, A.A. A multi-platform metabolomics approach demonstrates changes in energy metabolism and the transsulfuration pathway in Chironomus tepperi following exposure to zinc. Aquat. Toxicol. 2015, 162, 54–65. [Google Scholar] [CrossRef]
- Aquilano, K.; Baldelli, S.; Ciriolo, M.R. Glutathione: New roles in redox signaling for an old antioxidant. Front. Pharmacol. 2014, 5, 196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McBean, G.J. The transsulfuration pathway: A source of cysteine for glutathione in astrocytes. Amino Acids 2012, 42, 199–205. [Google Scholar] [CrossRef] [Green Version]
- Jozefczak, M.; Remans, T.; Vangronsveld, J.; Cuypers, A. Glutathione is a key player in metal-induced oxidative stress defenses. Int. J. Mol. Sci. 2012, 13, 3145–3175. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Ballatori, N. Endogenous glutathione conjugates: Occurrence and biological functions. Pharmacol. Rev. 1998, 50, 335–356. [Google Scholar]
- Rabenstein, D.L. Metal complexes of glutathione and their biological significance. In Glutathione: Chemical Biochemical and Medical Aspects: Coenzymes and Cofactors; Dolphin, D., Auramovibc, O., Poulson, R., Eds.; Wiley: New York, NY, USA, 1989; Volume 3, pp. 147–186. [Google Scholar]
- Maracine, M.; Segner, H. Cytotoxicity of metals in isolated fish cells: Importance of the cellular glutathione status. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 1998, 120, 83–88. [Google Scholar] [CrossRef]
- Lu, S.C. Regulation of glutathione synthesis. Mol. Asp. Med. 2009, 30, 42–59. [Google Scholar] [CrossRef] [Green Version]
- Marí, M.; Morales, A.; Colell, A.; García-Ruiz, C.; Fernández-Checa, J.C. Mitochondrial glutathione, a key survival antioxidant. Antioxid. Redox Signal. 2009, 11, 2685–2700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geret, F.; Serafim, A.; Barreira, L.; Bebianno, M.J. Response of antioxidant systems to copper in the gills of the clam Ruditapes decussatus. Mar. Environ. Res. 2002, 54, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Bigot, A.; Vasseur, P.; Rodius, F. SOD and CAT cDNA cloning, and expression pattern of detoxification genes in the freshwater bivalve Unio tumidus transplanted into the Moselle river. Ecotoxicology 2010, 19, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Maria, V.L.; Bebianno, M.J. Antioxidant and lipid peroxidation responses in Mytilus galloprovincialis exposed to mixtures of benzo(a)pyrene and copper. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2011, 154, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Atli, G.; Grosell, M. Characterization and response of antioxidant systems in the tissues of the freshwater pond snail (Lymnaea stagnalis) during acute copper exposure. Aquat. Toxicol. 2016, 176, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Qiu, L.; Wu, H.; Liu, X.; You, L.; Pei, D.; Chen, L.; Wang, Q.; Zhao, J. Expression profiles of seven glutathione S-transferase (GST) genes from Venerupis philippinarum exposed to heavy metals and benzo[a]pyrene. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2012, 155, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Gireesh, R.; Biju, A.; Muthiah, P. Biochemical changes during larval development in the short neck clam, Paphia malabarica Chemnitz. Aquac. Res. 2009, 40, 1510–1515. [Google Scholar] [CrossRef] [Green Version]
- Matias, D.; Joaquim, S.; Ramos, M.; Sobral, P.; Leitão, A. Biochemical compounds’ dynamics during larval development of the carpet-shell clam Ruditapes decussatus (Linnaeus, 1758): Effects of mono-specific diets and starvation. Helgol. Mar. Res. 2010, 65, 369–379. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Lazo, C.; Martínez-Pita, I. Biochemical and energy dynamics during larval development of the mussel Mytilus galloprovincialis (Lamarck, 1819). Aquaculture 2012, 358, 71–78. [Google Scholar] [CrossRef]
- Liu, H.; Guo, Z.; Zheng, H.; Wang, S.; Wang, Y.; Liu, W.; Zhang, G. Functional characterization of a Δ5-like fatty acyl desaturase and its expression during early embryogenesis in the noble scallop Chlamys nobilis Reeve. Mol. Biol. Rep. 2014, 41, 7437–7445. [Google Scholar] [CrossRef]
- Da Costa, F.; Robert, R.; Quéré, C.; Wikfors, G.H.; Soudant, P. Essential fatty acid assimilation and synthesis in larvae of the bivalve Crassostrea gigas. Lipids 2015, 50, 503–511. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, A.K.; Turchini, G.M.; Jahangard, S.; Ingram, B.A.; Sherman, C.D. Effects of different dietary microalgae on survival, growth, settlement and fatty acid composition of blue mussel (Mytilus galloprovincialis) larvae. Aquaculture 2010, 309, 115–124. [Google Scholar] [CrossRef]
- Paolisso, G.; Gambardella, A.; Tagliamonte, M.R.; Saccomanno, F.; Salvatore, T.; Gualdiero, P.; D’Onofrio, M.V.; Howard, B.V. Does free fatty acid infusion impair insulin action also through an increase in oxidative stress? J. Clin. Endocrinol. Metab. 1996, 81, 4244–4248. [Google Scholar] [CrossRef]
- Tripathy, D.; Aljada, A.; Dandona, P. Free fatty acids (FFA) and endothelial dysfunction; Role of increased oxidative stress and inflammation. Diabetologia 2003, 46, 300–301. [Google Scholar] [CrossRef] [Green Version]
- Tripathy, D.; Mohanty, P.; Dhindsa, S.; Syed, T.; Ghanim, H.; Aljada, A.; Dandona, P. Elevation of free fatty acids induces inflammation and impairs vascular reactivity in healthy subjects. Diabetes 2003, 52, 2882–2887. [Google Scholar] [CrossRef] [Green Version]
- Morgan, M.J.; Liu, Z.G. Crosstalk of reactive oxygen species and NF-κB signaling. Cell Res. 2011, 21, 103–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Wang, M.; Zhao, J.; Wang, L.; Gao, Y.; Zhang, H.; Song, L. The increased transcriptional response and translocation of a Rel/NF-κB homologue in scallop Chlamys farreri during the immune stimulation. Fish Shellfish Immunol. 2013, 34, 1209–1215. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhang, R.; Zhang, L.; Zou, J.; Xing, Q.; Dou, H.; Hu, X.; Zhang, L.; Wang, R.; Bao, Z. Characterizations and expression analyses of NF-κB and Rel genes in the Yesso scallop (Patinopecten yessoensis) suggest specific response patterns against Gram-negative infection in bivalves. Fish Shellfish Immunol. 2015, 44, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Baker, R.G.; Hayden, M.S.; Ghosh, S. NF-κB, inflammation, and metabolic disease. Cell Metab. 2011, 13, 11–22. [Google Scholar] [CrossRef] [Green Version]
- Janssen-Heininger, Y.M.; Poynter, M.E.; Baeuerle, P.A. Recent advances towards understanding redox mechanisms in the activation of nuclear factor κb. Free Radic. Biol. Med. 2000, 28, 1317–1327. [Google Scholar] [CrossRef]
- Biswas, D.K.; Shi, Q.; Baily, S.; Strickland, I.; Ghosh, S.; Pardee, A.B.; Iglehart, J.D. NF-κB activation in human breast cancer specimens and its role in cell proliferation and apoptosis. Proc. Natl. Acad. Sci. USA 2004, 101, 10137–10142. [Google Scholar] [CrossRef]
- Hayden, M.S.; Ghosh, S. NF-κB in immunobiology. Cell Res. 2011, 21, 223–244. [Google Scholar] [CrossRef] [Green Version]
- Gilmore, T.D.; Wolenski, F.S. NF-κB: Where did it come from and why? Immunol. Rev. 2012, 246, 14–35. [Google Scholar] [CrossRef]
- Persichini, T.; Percario, Z.; Mazzon, E.; Colasanti, M.; Cuzzocrea, S.; Musci, G. Copper activates the NF-κB pathway in vivo. Antioxid. Redox Signal. 2006, 8, 1897–1904. [Google Scholar] [CrossRef]
- Korashy, H.M.; El-Kadi, A.O. The role of redox-sensitive transcription factors NF-κB and AP-1 in the modulation of the Cyp1a1 gene by mercury, lead, and copper. Free Radic. Biol. Med. 2008, 44, 795–806. [Google Scholar] [CrossRef]
- Ji, C.; Wang, Q.; Wu, H.; Tan, Q.; Wang, W.-X. A metabolomic study on the biological effects of metal pollutions in oysters Crassostrea sikamea. Mar. Pollut. Bull. 2016, 102, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.; Yu, D.; Wang, Q.; Li, F.; Zhao, J.; Wu, H. Impact of metal pollution on shrimp Crangon affinis by NMR-based metabolomics. Mar. Pollut. Bull. 2016, 106, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Ji, C.; Zhao, J.; Wu, H. Metabolic responses to metal pollution in shrimp Crangon affinis from the sites along the Laizhou Bay in the Bohai Sea. Mar. Pollut. Bull. 2016, 113, 536–541. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S. Malonate metabolism: Biochemistry, molecular biology, physiology, and industrial application. BMB Rep. 2002, 35, 443–451. [Google Scholar] [CrossRef] [Green Version]
- Wojtovich, A.P.; Brookes, P.S. The endogenous mitochondrial complex II inhibitor malonate regulates mitochondrial ATP-sensitive potassium channels: Implications for ischemic preconditioning. Biochim. Biophys. Acta Bioenerg. 2008, 1777, 882–889. [Google Scholar] [CrossRef] [Green Version]
- Döbrössy, M.D.; Büchele, F.; Nikkhah, G. Excitotoxic lesions of the rodent striatum. In Animal Models of Movement Disorders: Volume II; Lane, E.L., Dunnett, S.B., Eds.; Humana Press: Totowa, NJ, USA; Springer: New York, NY, USA, 2011; pp. 21–35. [Google Scholar]
- Fernandez-Gomez, F.J.; Galindo, M.F.; Gómez-Lázaro, M.; Yuste, V.J.; Comella, J.X.; Aguirre, N.; Jordán, J. Malonate induces cell death via mitochondrial potential collapse and delayed swelling through an ROS-dependent pathway. Br. J. Pharmacol. 2005, 144, 528–537. [Google Scholar] [CrossRef] [Green Version]
- De Oliveira Marques, F.; Hagen, M.E.K.; Pederzolli, C.D.; Sgaravatti, A.M.; Durigon, K.; Testa, C.G.; Wannmacher, C.M.D.; Wyse, A.T.D.S.; Wajner, M.; Dutra-Filho, C.S. Glutaric acid induces oxidative stress in brain of young rats. Brain Res. 2003, 964, 153–158. [Google Scholar] [CrossRef]
- Fighera, M.R.; Royes, L.F.F.; Furian, A.F.; Oliveira, M.S.; Fiorenza, N.G.; Frussa-Filho, R.; Petry, J.C.; Coelho, R.C.; Mello, C.F. GM1 ganglioside prevents seizures, Na+, K+-ATPase activity inhibition and oxidative stress induced by glutaric acid and pentylenetetrazole. Neurobiol. Dis. 2006, 22, 611–623. [Google Scholar] [CrossRef]
- Latini, A.; Ferreira, G.C.; Scussiato, K.; Schuck, P.F.; Solano, A.F.; Dutra-Filho, C.S.; Vargas, C.R.; Wajner, M. Induction of oxidative stress by chronic and acute glutaric acid administration to rats. Cell. Mol. Neurobiol. 2007, 27, 423–438. [Google Scholar] [CrossRef]
- Tian, F.; Fu, X.; Gao, J.; Ying, Y.; Hou, L.; Liang, Y.; Ning, Q.; Luo, X. Glutaric acid-mediated apoptosis in primary striatal neurons. BioMed Res. Int. 2014, 2014, 484731. [Google Scholar] [CrossRef] [Green Version]
- Soga, T.; Baran, R.; Suematsu, M.; Ueno, Y.; Ikeda, S.; Sakurakawa, T.; Kakazu, Y.; Ishikawa, T.; Robert, M.; Nishioka, T.; et al. Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J. Biol. Chem. 2006, 281, 16768–16776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujii, J.; Ito, J.I.; Zhang, X.; Kurahashi, T. Unveiling the roles of the glutathione redox system in vivo by analyzing genetically modified mice. J. Clin. Biochem. Nutr. 2011, 49, 70–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dello, S.A.; Neis, E.P.; de Jong, M.C.; van Eijk, H.M.; Kicken, C.H.; Damink, S.W.O.; Dejong, C.H. Systematic review of ophthalmate as a novel biomarker of hepatic glutathione depletion. Clin. Nutr. 2013, 32, 325–330. [Google Scholar] [CrossRef] [PubMed]



 ) indicate metabolite fold changes in Cu-treated embryos relative to control embryos (green circles;
) indicate metabolite fold changes in Cu-treated embryos relative to control embryos (green circles;  ) plotted using a log2 scale.
) plotted using a log2 scale.
 ) indicate metabolite fold changes in Cu-treated embryos relative to control embryos (green circles;
) indicate metabolite fold changes in Cu-treated embryos relative to control embryos (green circles;  ) plotted using a log2 scale.
) plotted using a log2 scale.





| Copper Species/Fraction | Concentration | Element | Concentration | Element | Concentration |
|---|---|---|---|---|---|
| Total dissolved Cu | 2.90 µg·L−1 | Aluminium | <13.0 µg·L−1 | Magnesium | 1.49 g·L−1 |
| Humic-bound Cu | 2.76 µg·L−1 | Arsenic | <4.2 µg·L−1 | Manganese | <1.1 µg·L−1 |
| Free Cu2+ | 0.01 µg·L−1 | Barium | 5.9 µg·L−1 | Mercury | <80.0 ng·L−1 |
| Inorganic-bound Cu | 0.123 µg·L−1 | Beryllium | <0.63 µg·L−1 | Molybdenum | 11.1 µg·L−1 |
| Bioavailable Cu | 0.133 µg·L−1 | Boron | 4.0 mg·L−1 | Nickel | <6.3 µg·L−1 |
| Nutrient Profile | Concentration | Bromine | 82.0 mg·L−1 | Phosphorus | 22.0 µg·L−1 |
| Total ammoniacal-N | <10.0 µg·L−1 | Cadmium | <0.21 µg·L−1 | Potassium | 0.4 g·L−1 |
| Nitrite-N | <2.0 µg·L−1 | Cesium | <1.9 µg·L−1 | Rubidium | 0.12 mg·L−1 |
| Nitrate-N | 4.0 µg·L−1 | Calcium | 0.42 g·L−1 | Selenium | <4.2 µg·L−1 |
| Nitrite-N + Nitrate-N | 4.0 µg·L−1 | Carbon | 24.0 mg·L−1 | Silver | <0.43 µg·L−1 |
| Dissolved reactive P | 8.0 µg·L−1 | Chloride | 19.0 g·L−1 | Sodium | 11.2 g·L−1 |
| Other | Concentration | Chromium | <1.1 µg·L−1 | Strontium | 8.7 mg·L−1 |
| Alkalinity (CaCO3) | 116.0 mg·L−1 | Cobalt | <0.63 µg·L−1 | Thallium | <0.21 µg·L−1 |
| Total Kjeldahl Nitrogen | <0.2 mg·L−1 | Fluoride | 1.6 mg·L−1 | Tin | <1.7 µg·L−1 |
| Reactive silica (SiO2) | 0.29 mg·L−1 | Iron | 5.4 µg·L−1 | Uranium | 3.3 µg·L−1 |
| Dissolved non-purgeable organic C | 1.1 mg·L−1 | Lead | <1.1 µg·L−1 | Vanadium | 1.9 µg·L−1 |
| Lithium | 0.19 mg·L−1 | Zinc | 6.3 µg·L−1 |
| Copper Species/Fraction | Experimental Copper Concentrations | |||
|---|---|---|---|---|
| Control | Low Dose | Medium Dose | High Dose | |
| Target total dissolved Cu | 0.0 µg·L−1 | 100 µg·L−1 | 200 µg·L−1 | 300 µg·L−1 |
| Measured total dissolved Cu | 2.6 µg·L−1 | 130 µg·L−1 | 250 µg·L−1 | 370 µg·L−1 |
| Humic-bound Cu | 26.0 ng·L−1 | 1.6 µg·L−1 | 9.0 µg·L−1 | 57.0 µg·L−1 |
| EDTA-bound Cu | 2.6 µg·L−1 | 130 µg·L−1 | 240 µg·L−1 | 260 µg·L−1 |
| Free Cu2+ | 0.06 ng·L−1 | 3.2 ng·L−1 | 83.0 ng·L−1 | 4.3 µg·L−1 |
| Inorganic-bound Cu | 4.0 ng·L−1 | 39.0 ng·L−1 | 0.98 µg·L−1 | 51.0 µg·L−1 |
| Non-bioavailable Cu | 2.6 µg·L−1 | 130 µg·L−1 | 249 µg·L−1 | 310 µg·L−1 |
| Bioavailable Cu | 0.47 ng·L−1 | 0.04 µg·L−1 | 1.1 µg·L−1 | 50.3 µg·L−1 |
| Biomarker | 0.04 µg·L−1 | 1.10 µg·L−1 | 50.3 µg·L−1 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 3 h | 18 h | 42 h | 72 h | 3 h | 18 h | 42 h | 72 h | 3 h | 18 h | |
| DNA damage | - | - | - | - | - | ↑ | ↑ | ↑ | ↑ | ↑ |
| Protein damage | - | - | - | - | - | ↑ | ↑ | ↑ | ↑ | ↑ |
| Lipid damage | ↓ | - | ↑ | - | - | ↑ | ↑ | ↑ | ↑ | ↑ |
| GSH | - | - | - | - | - | - | - | - | ↓ | ↓ |
| CAT | - | - | - | - | - | - | ↑ | ↑ | ↓ | ↓ |
| SOD | - | - | - | - | - | - | ↑ | ↑ | ↓ | ↓ |
| GR | - | - | - | - | - | - | ↑ | ↑ | ↓ | ↓ |
| GST | - | - | - | - | ↑ | - | ↑ | ↑ | - | ↓ |
| GPx | - | - | - | - | ↑ | ↑ | ↑ | ↑ | - | ↓ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Young, T.; Gale, S.L.; Ragg, N.L.C.; Sander, S.G.; Burritt, D.J.; Benedict, B.; Le, D.V.; Villas-Bôas, S.G.; Alfaro, A.C. Metabolic Regulation of Copper Toxicity during Marine Mussel Embryogenesis. Metabolites 2023, 13, 838. https://doi.org/10.3390/metabo13070838
Young T, Gale SL, Ragg NLC, Sander SG, Burritt DJ, Benedict B, Le DV, Villas-Bôas SG, Alfaro AC. Metabolic Regulation of Copper Toxicity during Marine Mussel Embryogenesis. Metabolites. 2023; 13(7):838. https://doi.org/10.3390/metabo13070838
Chicago/Turabian StyleYoung, Tim, Samantha L. Gale, Norman L. C. Ragg, Sylvia G. Sander, David J. Burritt, Billy Benedict, Dung V. Le, Silas G. Villas-Bôas, and Andrea C. Alfaro. 2023. "Metabolic Regulation of Copper Toxicity during Marine Mussel Embryogenesis" Metabolites 13, no. 7: 838. https://doi.org/10.3390/metabo13070838