- 1MARUM – Center for Marine Environmental Sciences, University of Bremen, Bremen, Germany
- 2Max Planck Institute for Marine Microbiology, Bremen, Germany
- 3Faculty of Geosciences, University of Bremen, Bremen, Germany
Consortia of anaerobic methanotrophic archaea (ANME) and sulfate-reducing bacteria mediate the anaerobic oxidation of methane (AOM) in marine sediments. However, even sediment-free cultures contain a substantial number of additional microorganisms not directly related to AOM. To track the heterotrophic activity of these community members and their possible relationship with AOM, we amended meso- (37°C) and thermophilic (50°C) AOM cultures (dominated by ANME-1 archaea and their partner bacteria of the Seep-SRB2 clade or Candidatus Desulfofervidus auxilii) with L-leucine-3-13C (13C-leu). Various microbial lipids incorporated the labeled carbon from this amino acid, independent of the presence of methane as an energy source, specifically bacterial fatty acids, such as iso and anteiso-branched C15:0 and C17:0, as well as unsaturated C18:1ω9 and C18:1ω7. In natural methane-rich environments, these bacterial fatty acids are strongly 13C-depleted. We, therefore, suggest that those fatty acids are produced by ancillary bacteria that grow on 13C-depleted necromass or cell exudates/lysates of the AOM core communities. Candidates that likely benefit from AOM biomass are heterotrophic bacterial members of the Spirochetes and Anaerolineae—known to produce abundant branched fatty acids and present in all the AOM enrichment cultures. For archaeal lipids, we observed minor 13C-incorporation, but still suggesting some 13C-leu anabolism. Based on their relatively high abundance in the culture, the most probable archaeal candidates are Bathyarchaeota, Thermoplasmatales, and Lokiarchaeota. The identified heterotrophic bacterial and archaeal ancillary members are likely key players in organic carbon recycling in anoxic marine sediments.
Introduction
Methane is the most abundant hydrocarbon in marine sediments. The emission of methane from sediments into the water column and eventually the atmosphere is attenuated by the anaerobic oxidation of methane (AOM), which is performed by anaerobic methane-oxidizing archaea (ANME) and sulfate-reducing bacteria consortia (SRB) (Hinrichs et al., 1999; Boetius et al., 2000; Reeburgh, 2007; Wegener et al., 2016). The ANMEs completely oxidize methane to carbon dioxide, and their partner bacteria use the reducing equivalents produced in this reaction for sulfate reduction (Orphan et al., 2001). This exchange likely involves direct electron transfer mediated by cytochromes and nanowires (McGlynn et al., 2015; Wegener et al., 2015). ANME archaea are found in three clades known as ANME-1, ANME-2, and ANME-3 (Orphan et al., 2002; Niemann et al., 2006). Psychro- and mesophilic ANMEs form a consortium with SRB of the Desulfosarcina/Desulfococcus (DSS), classified as Seep-SRB1, or Desulfobulbus group (Boetius et al., 2000; Michaelis et al., 2002; Niemann et al., 2006). The thermophilic ANME-1 archaea form a consortium with Candidatus Desulfofervidus auxilii (Ca. D. auxilii) in heated sediments (e.g., Guaymas Basin). In contrast to SRBs in the psychro- and mesophilic consortia, Ca. D. auxilii has been isolated using molecular hydrogen as an alternative electron donor (Krukenberg et al., 2016).
ANMEs are characterized by diagnostic lipid biomarker patterns. ANME-1 archaea predominantly synthesize glycerol dialkyl glycerol tetraethers (GDGTs) as opposed to ANME-2 and ANME-3 that produce archaeol-based diethers, predominantly hydroxyarchaeol (Blumenberg et al., 2004; Rossel et al., 2008, 2011). Nonetheless, a thermophilic ANME-1 AOM enrichment from the Guaymas Basin revealed a substantial quantity of archaeol lipids in comparison to GDGTs, especially in the active growth phase (Kellermann et al., 2016; Wegener et al., 2016). The corresponding lipid patterns of SRB partners determined from AOM environments and cultures are more diverse and taxonomically only partly distinctive (e.g., Hinrichs et al., 2000; Elvert et al., 2003, 2005; Blumenberg et al., 2004; Niemann and Elvert, 2008). It has been shown that bacterial fatty acids (FAs) from environments dominated by ANME-2 include large proportions of C16:1ω5c and cyclopropane (cy)-C17:0ω5,6 (Elvert et al., 2003; Blumenberg et al., 2004), while those dominated by ANME-1 predominantly produce ai-C15:0 (Blumenberg et al., 2004; Elvert et al., 2005). All of the aforementioned archaeal or bacterial lipids show strong 13C-depletions with δ13C values of –70‰ and lower, which are assumed to be caused by the distinctively low δ13C values of methane (e.g., Hinrichs et al., 1999; Thiel et al., 1999; Pancost et al., 2001; Orphan et al., 2002; Elvert et al., 2003; Blumenberg et al., 2004).
Multiple stable isotope probing (SIP) experiments indicate that ANMEs and their direct SRB partners predominantly assimilate inorganic carbon (Wegener et al., 2008; Kellermann et al., 2012). Specifically, ANME-1 was classified as a methane-oxidizing chemoorganoautotroph (Kellermann et al., 2012). Here, we used long-term meso- and thermophilic AOM enrichment cultures obtained from hydrocarbon-rich heated sediments in the Guaymas Basin. The mesophilic culture grown at 37°C (AOM37) is dominated by ANME-1 and Seep-SRB2; the thermophilic culture maintained at 50°C (AOM50) is dominated by ANME-1 and Ca. D. auxilii (Krukenberg et al., 2016; Wegener et al., 2016). Although maintained for 5 years with methane as the sole energy source, these cultures contain substantial numbers of additional bacteria and archaea (Wegener et al., 2016). The functions and carbon sources of these ancillary microbes and their relationship with the AOM consortia remain largely unknown. Kellermann et al. (2012) suggested that these uncultured microbes may be heterotrophs, which likely feed on labile organic compounds, such as acetate or protein-like dissolved organic carbon detected in the pore waters of AOM environments (Heuer et al., 2006; Yoshinaga et al., 2015; Yang et al., 2020; Hu et al., 2021; Pérez Castro et al., 2021).
Leucine is one of the most abundant amino acids produced by microorganisms and, if released into the environment, becomes a carbon, nitrogen, and energy source (Kirchman et al., 1985). Because leucine metabolism was found to be particularly essential during starvation conditions (Harwood and Canale-Parola, 1981; Mårdén et al., 1987), it is ideal for tracking heterotrophic activity in slow-growing enrichment cultures, such as AOM consortia. To explore the activity of these heterotrophic community members and their signaling lipids in AOM environments, we incubated active Guaymas Basin AOM enrichment cultures with 13C-leu, a particular precursor for iso-branched FAs (cf. Aepfler et al., 2019), which are abundant in natural ANME-1 systems. Additionally, we used the same cultures devoid of methane to suppress the activity of AOM consortia members and to track the utilization of leucine for lipid biosynthesis by non-AOM microbes. Based on our 13C-leu incubation and published microbial community data on the same cultures (Wegener et al., 2016; Krukenberg et al., 2018), we were able to trace ancillary heterotrophic bacteria and archaea in AOM enrichment cultures, detected by strong 13C-enrichments of diagnostic FAs but only minor for archaeal lipids, highlighting the identification of branched fatty acids as indicators of bacterial heterotrophy.
Materials and Methods
Anaerobic Oxidation of Methane Cultures
The production and maintenance of the sediment-free AOM cultures from the Guaymas Basin were performed as described before (Wegener et al., 2016; Laso-Pérez et al., 2018). In brief, both AOM37 and AOM50 were incubated with marine sulfate reducer medium supplemented with trace amounts of vitamins (Widdel and Bak, 1992) under a CH4:CO2 atmosphere (2.5 atm; 90:10) at temperatures of 37°C and 50°C, respectively. The initial concentration of sulfate was 28 mM. The carbon isotopic composition of methane used was −35‰ (Wegener et al., 2021), and sulfide concentrations were measured as described before (Cord-Ruwisch, 1985). When sulfide concentrations exceeded 15 mM, microbial biomass was transferred into a fresh medium. Under these conditions, the AOM37 and AOM50 cultures show doubling times of 69 and 55 days, respectively (Holler et al., 2011). The sulfate reducer Ca. D. auxilii was isolated from AOM50 with hydrogen as the sole electron donor and sulfate as an electron acceptor. It is chemolithoautotrophic and grows at temperatures between 50 and 70°C, and has a doubling time of 4–6 days (Krukenberg et al., 2016).
Experimental Setup
For all experiments with the AOM cultures, the culture medium was exchanged, and cultures were equally distributed in 156 ml cultivation bottles. In the case of Ca. D. auxilii, new dilutions were prepared (5 ml of active culture for inoculation). 13C-leu was dissolved in Milli-Q water and sterilized by filtration (Minisart High Flow, PES, 28 mm, 0.1 μm, sterile). The AOM37 and AOM50 cultures were amended with 100 μM of sterilized 13C-leu and incubated under different experimental conditions for 28 days (Table 1): experiment 1 with CH4 and 13C-leu to track characteristic lipid production by microbial community members involved in leucine metabolism during active AOM; experiment 2 with CH4 and without 13C-leu as a negative control; experiment 3 with 13C-leu and without CH4 as a positive control to specifically track ancillary community members and identify their lipids by suppressing the activity of AOM consortia. Through these three experiments, we were able to target ancillary microbial communities existing in the current AOM cultures and constrain their potential heterotrophic capabilities. In contrast, experiment 4 utilized the autotrophic Ca. D. auxilii culture and was likewise amended with 100 μM of sterilized 13C-leu. This experiment lasted for 40 days, and it thoroughly tested whether the partner bacterium Ca. D. auxilii can metabolize 13C-leu and constrain its lipid pattern. A 13C-leu concentration of 100 μM was chosen to ensure a sufficient supply of substrate and to maximize the potential to observe various pathways of leucine metabolism during prolonged incubation, even though the concentration is higher than existing leucine data from estuarine pore water (up to 3 μM, Henrichs and Farrington, 1979).
Determination of Sulfide Concentration and Isotopic Composition of Dissolved Inorganic Carbon
Sulfide concentrations were used to monitor the growth of AOM consortia and Ca. D. auxilii. The medium subsampling of AOM37 for sulfide concentrations analysis was at 0, 0.5, 3, 7, 14, 21, and 28 days; AOM50 medium subsampling was at 0, 7, 21, and 28 days; and Ca. D. auxilii medium subsampling was at 0 and 40 days. The subsampling for the measurements of the carbon isotopic composition of dissolved inorganic carbon (δ13CDIC) was performed on the same days to constrain the leucine mineralization (Aepfler et al., 2019). In brief, 1 ml of the sample was taken by syringe from the incubation serum bottles and filtered through a 0.2 μm filter (Minisart regenerated cellulose syringe filter, 15 mm) to remove cells and other particles. Finally, samples were acidified with 100 μl phosphoric acid overnight in an Exetainer vial pre-purged with CO2-free air before isotopic analysis. All samples were measured with a Thermo Scientific Delta Ray isotope ratio infrared spectrometer with an analytical error of ±1‰, which is obtained by repeated measurement of the laboratory CO2 reference gas (n = 8). All isotopic values are reported in the delta notation as δ13C relative to the Vienna PeeDee Belemnite (VPDB) standard.
Lipid Extraction, Identification, Quantification, and Isotopic Analysis
Due to potential contamination, we avoided subsampling for lipid analysis from the same bottle as used for sulfide concentration and δ13CDIC determination by obtaining biomass from replicate samples. Cell pellets from these incubations were extracted wet using a modified Bligh and Dyer protocol (Sturt et al., 2004). Before extraction, 1 μg of 1,2-diheneicosanoyl-sn-glycero-3-phosphocholine and 2-methyloctadecanoic acid were added as internal standards. Polar lipid-derived fatty acids (PLFAs) in the total lipid extract (TLE) were converted to fatty acid methyl esters (FAMEs) using saponification with KOH/MeOH and derivatization with BF3/MeOH (Elvert et al., 2003). Archaeal intact polar lipids (IPLs) in the TLE were separated from the apolar core lipids (CLs) using preparative liquid chromatography (Meador et al., 2015), followed by ether cleavage of both fractions with BBr3 in dichloromethane and reduction of the resulting alkyl bromides with superhydride to form isoprenoid hydrocarbons (Jahn et al., 2004). The hydrocarbon products were purified by silica gel column chromatography using 4 ml of hexane as an eluent. Both FAMEs and isoprenoid hydrocarbons were measured by gas chromatography coupled to flame ionization detection (GC-FID, Thermo Finnigan Trace GC) for quantification and gas chromatography-mass spectrometry (GC-MS, Trace GC coupled to Trace MS, both from Thermo Finnigan) for structural identification using the protocols described by Aepfler et al. (2019). Using the same GC conditions, lipid δ13C values were determined by GC-isotope ratio-MS (Thermo Finnigan Trace GC coupled to a Thermo Scientific Delta V Plus) connected via a GC IsoLink interface and are reported relative to VPDB. The precision of a lab FA standard (2-methyloctadecanoic acid, n = 3) was greater than 0.7‰, while the deviations of duplicate isotopic measurement of sample FAs were between ± 1‰ and ± 100‰ (for PLFAs with label uptake of >1,000‰).
The incorporation of 13C-leu into bacterial lipids expressed as a percentage of 13C incorporation was calculated as the product of excess 13C and the amount of FA carbon based on the quantification via GC-FID. Excess 13C is derived from the difference between the fractional abundance (F) of 13C in FAs after 28 days relative to T0 with F = 13C/(13C + 12C) = R/(R + 1) and R being derived from the measured δ13C values as R = (δ13C/1,000 + 1) × RVPDB.
Results
Contents and δ13C Values of Microbial Lipids in the Original Cultures
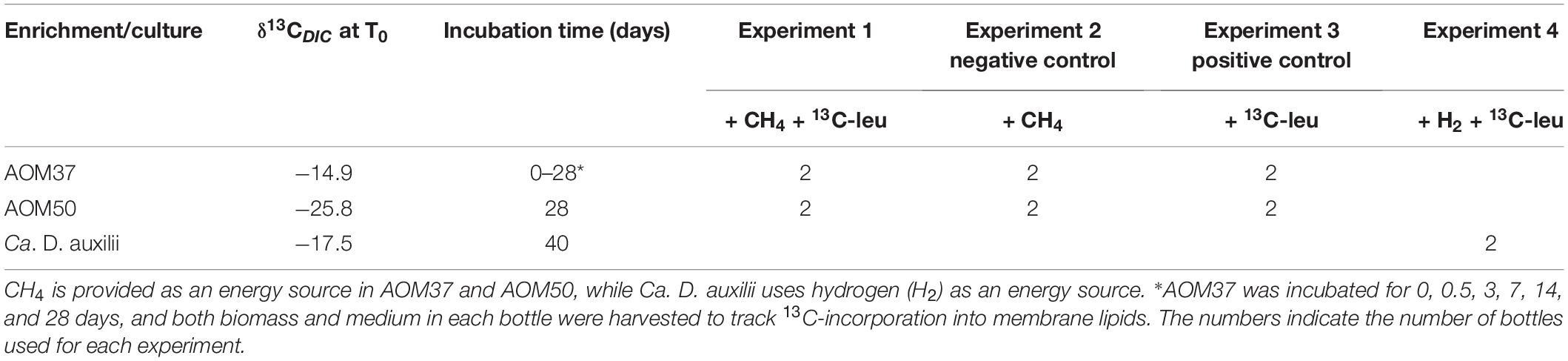
The FA distribution in the original AOM37 culture mainly consisted of C18:1ω7 (37%), C16:0 (24%), and C18:0 (21%) (Figure 1A). Branched-chain FAs accounted for 7% of the total. The AOM50 culture was dominated by C16:0 and C18.0 with a content of 46 and 28%, respectively. Branched-chain FAs accounted for 12%. The original Ca. D. auxilii culture had a FA pattern similar to AOM50, with C16:0 (40%) and C18:0 (52%) as the dominant FAs. Branched-chain FAs were below the detection limit in the Ca. D. auxilii culture. For archaea, we reported the relative content of phytane (Phy) and the three biphytanes (BP0, BP1, and BP2) derived from archaeols and GDGTs, respectively (Figure 1B). The content of Phy in AOM 37 was 23%, higher than that in AOM50 (10%). BPs had similar content in AOM37, with BP1 being highest at 30%. At the higher incubation temperature in AOM50, the BP pattern strongly shifted to BP2 (64%).
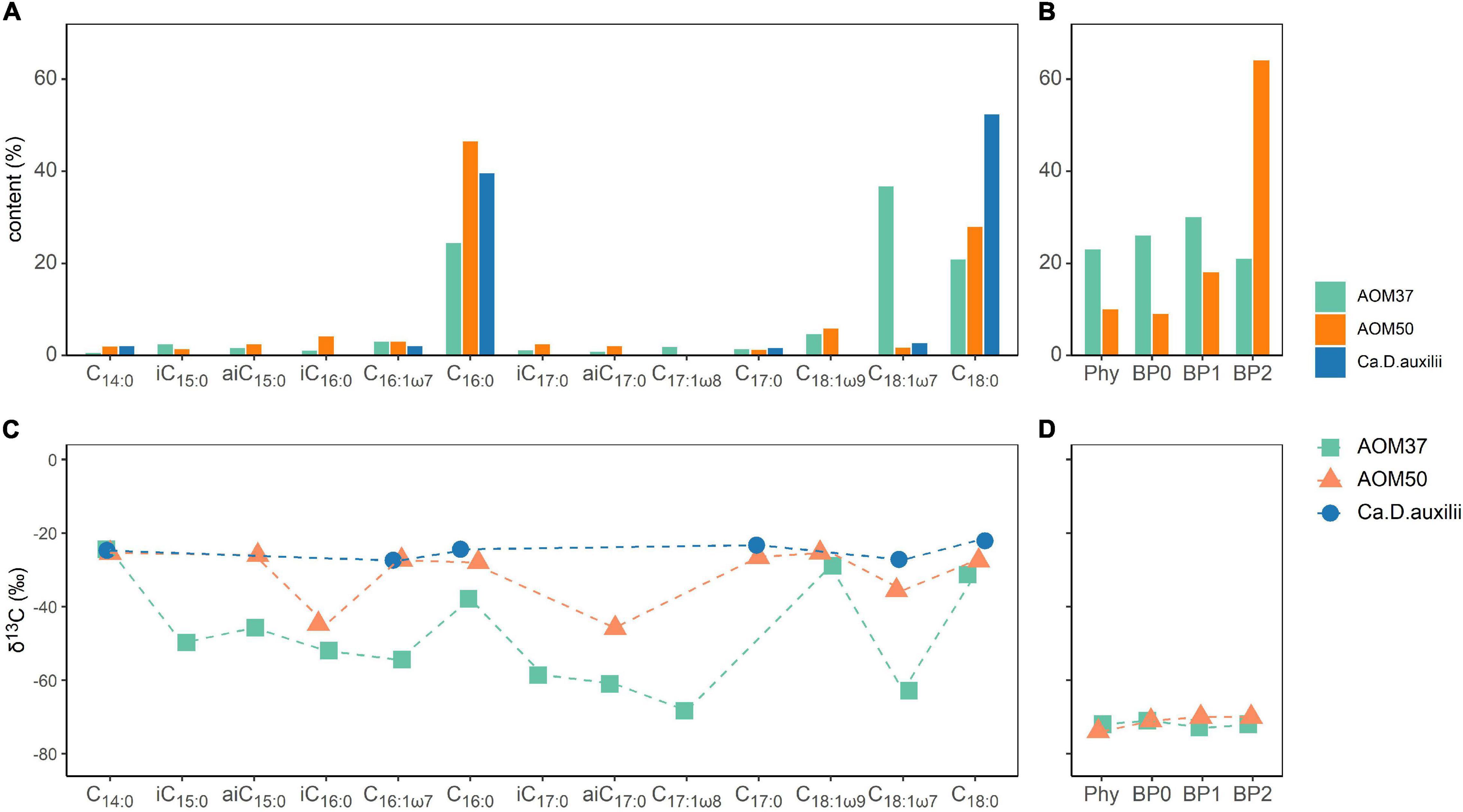
Figure 1. Bacterial and archaeal lipid patterns (A,B) and their δ13C values (C,D) in the original cultures AOM37 and AOM50 and the isolated bacterium Ca. D. auxilii. Phy and BP data are derived from total lipid extracts.
In AOM37, the δ13C values of monounsaturated FAs ranged from −55 to −68‰ except C18:1ω9 with a δ13C value of −29‰. Saturated C14:0, C16:0, and C18:0 FAs had less negative δ13C values between −25 and −38‰ (Figure 1C). The branched-chain FAs were more 13C-depleted, with δ13C values ranging from −46 to −61‰. FAs in AOM50 are generally less depleted in 13C than AOM37 and showed δ13C values between −25 and −45‰, with the most negative δ13C values found for the branched-chain FAs iC16:0 and aiC17:0. Different carbon fixation pathways of the respective partner bacterium may cause the difference in δ13C values of FAs in AOM37 and AOM50. The Seep-SRB-2 partner fixes carbon via the Wood-Ljungdahl pathway with a fractionation up to 36‰ (Preuß et al., 1989; Krukenberg et al., 2018), whereas Ca. D. auxilii uses the rTCA pathway with a lower carbon isotope fractionation of up to 12‰ (Wirsen et al., 2002; Suzuki et al., 2005; Krukenberg et al., 2016). The δ13C values of FAs in the culture of Ca. D. auxilii were even more positive, ranging between −21 and −28‰. δ13C values of TLE-derived Phy and BPs in the AOM37 and AOM50 were similar and around −70‰ (Figure 1D).
Temporal Development of Sulfide Production and δ13CDIC Values During Incubation
Sulfide concentrations (HS–) were measured to monitor the metabolic activity of the microorganisms involved in AOM and of Ca. D. auxilii (Figure 2A). In AOM37 and AOM50 cultures without CH4 (experiment 3), HS– concentrations remained stable, indicating a lack of methane-dependent sulfate reduction. When CH4 was provided (experiments 1 and 2), HS– increased gradually from 2.8 to 15.5 mM (ΔHS– = 12.7 mM) for AOM37 and from 2.2 to 24.7 mM (ΔHS– = 22.5 mM) for AOM50 within 28 days of incubation. There was no substantial difference between incubations with and without 13C-leu addition, indicating that leucine did not affect sulfate reduction. For Ca. D. auxilii, HS– increased from 2.7 to 26.1 mM (ΔHS– = 23.4 mM) after 40 days of incubation with hydrogen and 13C-leu (experiment 4).
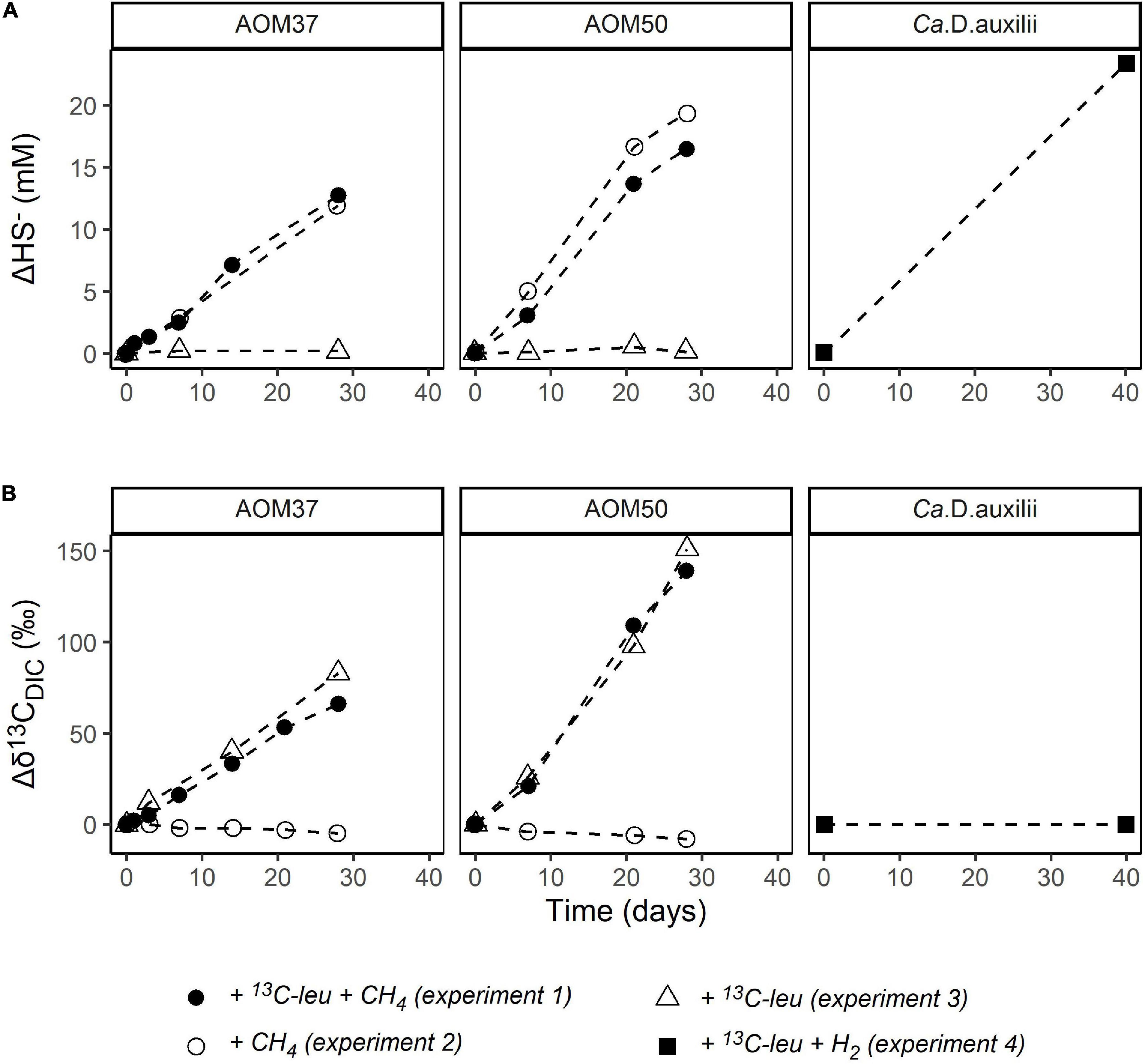
Figure 2. Development of ΔHS– (mM, A) and Δδ13CDIC (‰, B) relative to T0 in the incubation experiments with the AOM37 and AOM50 cultures over 28 days and the Ca. D. auxilii culture over 40 days.
We also measured the development of δ13CDIC values as an indicator of microbial oxidation of 13C-leu (Figure 2B). In AOM37 and AOM50 incubated with CH4 (experiment 2), δ13CDIC values decreased from −15 to −20‰ (Δδ13CDIC = −5‰) and −26 to −34‰ (Δδ13CDIC = −8‰), respectively, after 28 days of incubation caused by the oxidation of CH4. When both CH4 and 13C-leu were supplied for 28 days (experiment 1), the δ13CDIC value increased from −15 to +51‰ (Δδ13CDIC = 66‰) in AOM37 and from −26 to +118‰ (Δδ13CDIC = 144‰) in AOM50. If only 13C-leu was provided (experiment 3), δ13CDIC increased slightly more from −15 to +68‰ (Δδ13CDIC = 83‰) and from −26 to +125‰ (Δδ13CDIC = 151‰) in AOM37 and AOM50, respectively. The slight offset in Δδ13CDIC values between experiments 1 and 3 results from the dilution of the DIC signal with DIC derived from the oxidation of unlabeled CH4 with a δ13C value of −35‰ in the former experiment. The continuous increase of δ13CDIC values suggested a replete supply of 13C-leu during the whole incubation process. During incubation of Ca. D. auxilii, the addition of 13C-leu (experiment 4) did not alter the δ13CDIC values.
Alteration of 13C Values of Microbial Lipids in 13C-Leu Treatments
The lipid compositions of cultures that received 13C-leu were similar to those of the original cultures, suggesting that the overall community was stable during the incubations (Figure 1A and Supplementary Table 1), which did not cover a full doubling time. However, the 13C-leu additions in experiments 1 and 3 strongly altered the isotopic compositions of bacterial FAs (Figure 3 and Supplementary Table 1). In the AOM37 experiment, the 13C-leu addition already resulted in the increase of δ13C values by up to 260‰ relative to T0 (Δδ13C = δ13CT – δ13CT0) in iso-branched FAs iC15:0 and iC17:0 after 0.5 days. After 28 days of incubation, the anteiso-branched aiC15:0 incorporated most of the 13C (δ13C = 2,800‰), while iC15:0 showed a lower value of 2,100‰. In the AOM50 incubation, 13C-incorporation was even more pronounced, and δ13C values reached up to 6,400‰ for iC15:0 and iC17:0 after 28 days. Next to the branched FAs, the monounsaturated FA C18:1ω9 was highly labeled in the AOM37 incubation with a δ13C value of 2,200‰, which was not the case in AOM50. In both AOM37 and AOM50 enrichment cultures, carbon-numbered saturated FAs were at least 13C-labeled. Their δ13C values remained lower than those of the DIC, suggesting that the autotrophic partner bacteria mostly synthesize these lipids (Figure 2B). Overall, the δ13C values of FAs during incubation without CH4 (experiment 3) show a similar 13C-labeling strength to those in the incubation with CH4 (experiment 1, Supplementary Table 1), indicating that the incorporation of 13C-leu is independent of AOM activity. In contrast, the 40-day incubation with 13C-leu did not affect the lipid isotopic composition of the isolated autotrophic SRB partner Ca. D. auxilii (Supplementary Table 1), confirming its autotrophic lifestyle.
Figure 3. Development of δ13C values (in ‰ relative to T0) of bacterial FAs during 13C-leu incubation of AOM37 and AOM50 with and without (w/o) CH4 over 28 days (experiments 1 and 3, respectively).
We calculated the relative FA 13C-incorporation pattern of the heterotrophic bacterial community members of both AOM enrichment cultures (Figure 4) based on the FA content (Figure 1A) and respective δ13C values (Supplementary Table 1) after 28 days. In AOM37, the strongest 13C-incorporation is observed for the monounsaturated FAs C18:1ω9 (30.0%) and C18:1ω7 (20.6%), followed by iC15:0 (16.8%), aiC15:0 (13.3%), and iC17:0 (7.5%). In AOM50, we observed the highest 13C-incorporation in FAs iC15:0 (39.6%) and iC17:0 (31.2%), followed by iC16:0 (9.0%), while even-numbered FAs (C16:0 and C18:0) show much less 13C-incorporation, despite, as a sum, being the dominant fatty acids in all incubations (Figure 1). These results are independent of whether CH4 was supplied to the enrichment cultures or not (Figure 4).
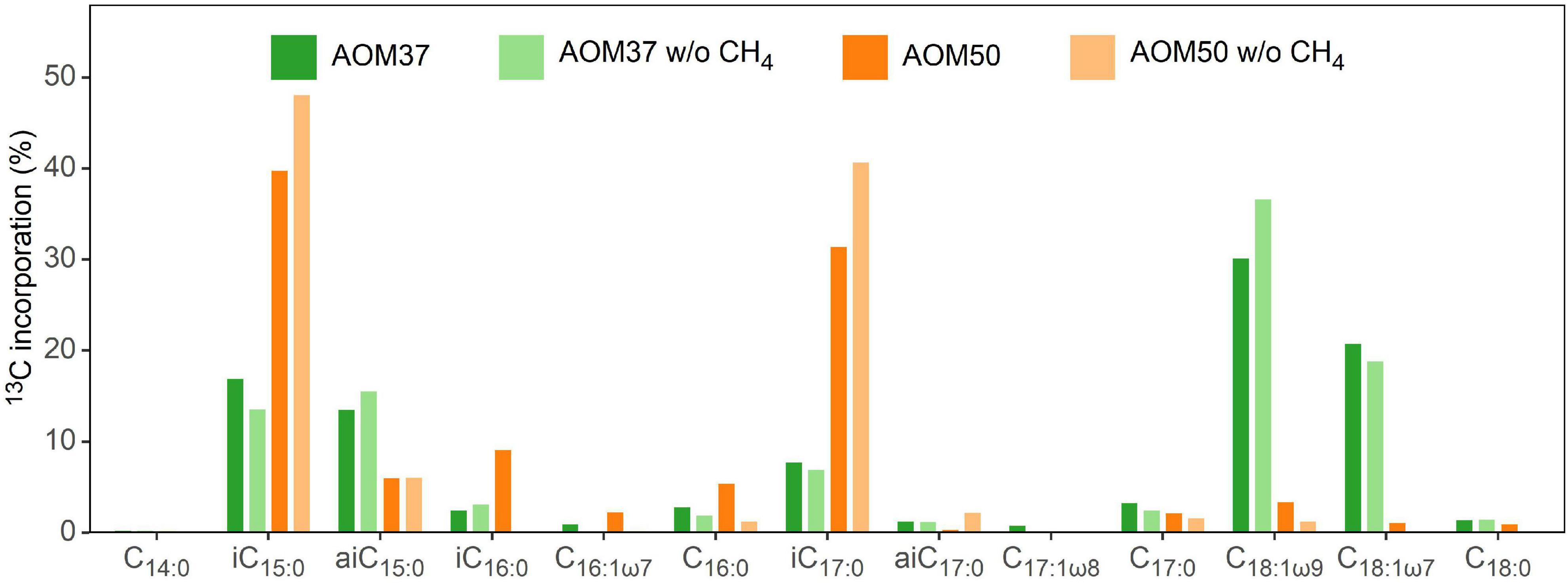
Figure 4. Pattern of 13C-incorporation into bacterial FAs in the AOM37 and AOM50 cultures during 13C-leu incubation with and without (w/o) CH4 after 28 days (experiments 1 and 3, respectively).
Changes in δ13C values relative to T0 of CL and IPL derived Phy and BPs in treatments with 13C-leu of AOM37 and AOM50 are shown in Figure 5. For all data on archaeal lipid-derived isoprenoid hydrocarbons, we refer to Supplementary Table 2. In both 13C-leu experiments, the 13C-incorporation into Phy and BPs was lower after 28 days (maximum δ13C value of 118‰ in IPL-Phy). These values are in the range of the corresponding 13C-label transfer into DIC (δ13CDIC values up to 151‰; Figure 2). However, the 13C-incorporation was independent of AOM activity. Throughout the experiments, BPs incorporated less 13C than Phy, regardless of whether they were being retrieved from the CL or IPL fractions. In particular, BP0, which is mostly derived from the GDGT caldarchaeol, had a δ13C value up to 16‰ higher in the 13C-leu-treated AOM37 and AOM50 culture than in the original cultures, independent of the addition of CH4 (experiments 1 and 3 compared to experiment 2). The CL-derived BP1 and BP2, AOM37, and AOM50 did not incorporate the 13C-label from 13C-leu (Supplementary Table 2). In contrast, CL- and IPL-derived Phy increased by up to 113‰ with CH4 (experiment 1) and up to 118‰ without CH4 (experiment 3) in AOM37 after 28 days. These values are specifically higher than δ13CDIC values, which increased by 66‰. For the AOM50 culture, minute amounts of IPL-derived Phy and BPs obtained after ether cleavage prevented isotope analyses.
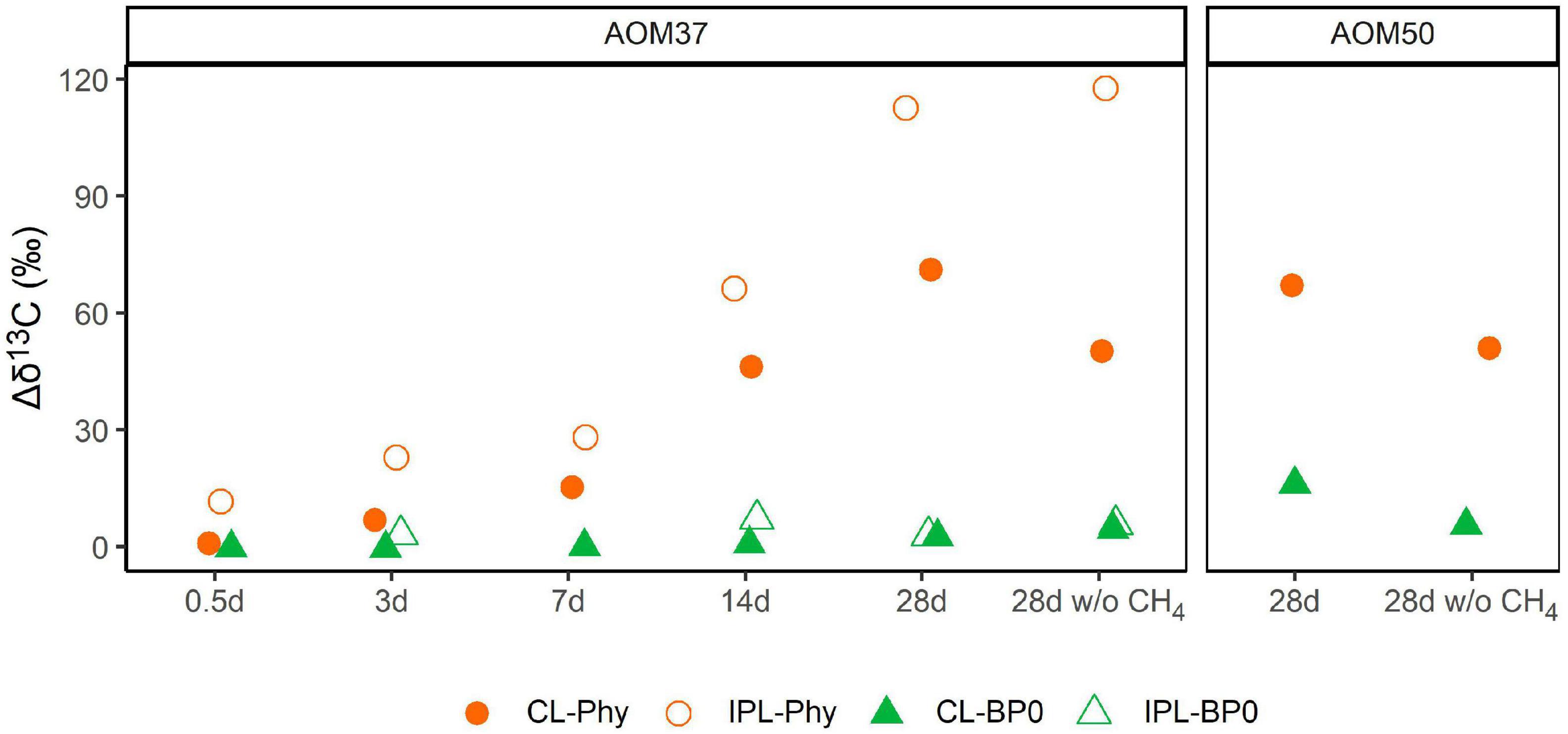
Figure 5. Development of δ13C values (in ‰ relative to T0) of archaeal lipid-derived isoprenoid hydrocarbons during 13C-leu incubation of AOM37 and AOM50 cultures with and without (w/o) CH4 over 28 days (experiments 1 and 3, respectively). In the AOM50 culture, the amounts of IPL-Phy and IPL-BP0 were too low to obtain δ13C values.
Discussion
Ancillary Microorganisms Grow on Leucine
In the AOM enrichment cultures, sulfide production quantitatively depends on CH4 as an energy source. The turnover of 13C-leu, as observed by changes in δ13CDIC values, had no measurable effect on sulfide production (Figure 2A) and occurred independent of the supply of CH4, indicating that leucine was predominantly metabolized by ancillary microbes not involved in AOM. This is in line with constant δ13CDIC values during the 40-day Ca. D. auxilii incubation (Figure 2B), providing concrete evidence that Ca. D. auxilii does not utilize leucine.
The microbial degradation and assimilation of leucine proceeds in diverse reactions. Leucine is deaminated and decarboxylated, resulting in isovaleryl-CoA, which can be used as a primer for odd-numbered iso-series FAs in bacteria, such as iC15:0 and iC17:0 (Kaneda, 1977; Aepfler et al., 2019). In addition, isovaleryl-CoA can be transformed via acetoacetate into acetyl-CoA (Yamauchi, 2010; Díaz-Pérez et al., 2016). Acetyl-CoA can either be completely oxidized or used for the synthesis of biomolecules, including the generation of bacterial FAs during elongation via malonyl-CoA or isoprenoid ether lipids in the case of halophilic archaea (Harwood and Canale-Parola, 1981; Yamada et al., 2006). Our experiments demonstrated the incorporation of 13C-leu into selected branched FAs, such as iC15:0 and iC17:0, independent of CH4 supply (Figure 4). Additionally, we observed a substantial 13C-incorporation into aiC15 and aiC17, which is explained by the production of a 2-methylbutyric acid intermediate during leucine catabolism under starvation conditions (Ganesan et al., 2006; Díaz-Pérez et al., 2016). This leads to the formation of 2-methylbutyryl-CoA, which serves as a primer molecule for the synthesis of anteiso FAs. Similarly, the interconversion of leucine and valine gives rise to isobutyryl-CoA (Monticello and Costilow, 1982), which serves as a primer of even-numbered iso-branched FAs, such as iC16. The high labeling of these fatty acids suggests that abundant heterotrophic bacteria not involved in sulfate-dependent AOM, such as Spirochetes or Anaerolineae (Supplementary Table 3), are generally active and likely to thrive on free amino acids or other protein-like organic matter (Hu et al., 2021). These carbon pools are highly 13C-depleted if derived from the biomass of AOM consortia in natural environments (Takano et al., 2018; Hu et al., 2021).
The partner bacterium Ca. D. auxilii neither incorporate 13C-leu into its dominant FAs C16:0 and C18:0, nor into any other FAs (Supplementary Table 1). The incorporation of 13C-leu differs between the meso- and thermophilic AOM enrichment cultures at 37°C and 50°C, respectively (Figures 3, 4). Compared to AOM37, AOM50 tends to channel more 13C from leucine into iso-branched than anteiso-branched FAs (AOM50, iso:anteiso = 92:8; AOM37, iso:anteiso = 63:37). Moreover, in AOM37, we observe a predominant 13C-incorporation into straight-chain FAs C18:1ω9 and C18:1ω7. These straight-chain FAs are likely synthesized by downstream 13C-leu products, such as acetate, and are probably derived from ancillary heterotrophic bacterial community members in AOM37 but not in AOM50, such as Spirochetes (Supplementary Table 3).
In the incubation of AOM37 and AOM50 with 13C-leu, Phy and BPs (derived from archaeal di- and tetraether lipids, respectively) incorporated much less 13C than bacterial FAs (Figures 3, 5). In the AOM37 culture, IPL-derived Phy provided a 47‰ stronger change in its δ13C value (Δδ13C = 113‰, Figure 5) than the corresponding DIC (Δδ13C = 66‰, Figure 2) after 28 days. This divergence suggests that archaeal IPLs are not solely biosynthesized via DIC assimilation in AOM37 (Supplementary Figure 1). Eventually, possible carbon substrates are leucine or, more likely, secondary metabolites, such as acetate, which is released during leucine catabolism of heterotrophic bacteria (Aepfler et al., 2019). Unfortunately, we could not examine this relationship in AOM50 due to very few isoprenoid hydrocarbons obtained after ether cleavage. In the 13C-leu incubations of our study, the relative 13C-enrichment of BPs was negligible compared to Phy but reached up to 16‰ for CL-derived BP0 in AOM50. Based on our short-time incubation, this is in agreement with former labeling experiments of ANME-1 dominated Guaymas Basin sediments using 13CDIC and D2O, which revealed an initial production of diether lipids that are later transformed into tetraether lipids (Kellermann et al., 2016). The observed isotopic evidence of the enhanced formation of 13C-enriched archaeal lipids over time suggests that some archaea participated in the leucine metabolism or the assimilation of metabolic intermediates. Nonetheless, their role in amino acid mineralization seems to be less important than heterotrophic bacterial community members due to their low 13C-label incorporation.
Minor Bacterial and Archaeal Community Members Thrive on Anaerobic Oxidation of Methane Necromass
Methane-rich sediments contain a large number of AOM consortia and diverse host archaeal and bacterial communities, with a substantial proportion of heterotrophic microorganisms (Biddle et al., 2006; Ruff et al., 2015; Dombrowski et al., 2018; Pérez Castro et al., 2021). These heterotrophs coexist with AOM consortia in natural environments and enrichment cultures, even after many years of maintenance (Wegener et al., 2016). Prior microbial composition analysis of the Guaymas Basin AOM cultures revealed that ANME-1 archaea and their partners dominate AOM37 and AOM50 (Seep-SRB2 and Ca. D. auxilii, Holler et al., 2011; Wegener et al., 2015, 2016). Ancillary microbial communities, identified by amplified 16S rRNA gene sequences of the AOM37 and the AOM50 culture (Wegener et al., 2016) and 16S rRNA genes recruited from the metagenomes of the AOM37 and the AOM60 culture (Supplementary Table 3; Krukenberg et al., 2018), are presumably unrelated to AOM. These include Anaerolineaceae and Spirochetes, and Candidate divisions JS1, WS3, and KB, many of which are known to be heterotrophs. Anaerolineae, which occupy up to 3.7% of the Guaymas Basin cultures (Supplementary Table 3), are strictly anaerobic heterotrophs and thrive on carbohydrates and amino acids (Rosenkranz et al., 2013; Liang et al., 2016). The cultured strains of this group produce mainly iso- and anteiso-C15 and C17 FAs (Yamada et al., 2006). Spirochetes similarly thrive on the degradation of carbohydrates and proteins (Paster, 2010; Dong et al., 2018) and are abundant in anoxic hydrocarbon-rich habitats (Dong et al., 2018). The AOM37 culture contains up to 5% Spirochetes, whereas these heterotrophs are absent in the AOM50 culture (Supplementary Table 3). Spirochetes primarily synthesize branched fatty acids, but some also produce substantial amounts of C18:1ω9 and C18:1ω7 (Livermore and Johnson, 1974; Vishnuvardhan Reddy et al., 2013). Hence, in the AOM37 culture, the substantial 13C-incorporation into branched fatty acids and C18:1ω9 and C18:1ω7 FAs is most likely due to the Spirochetes activity (Figure 4). The production of the latter can be attributed to the prolonged transformation and oxidation of isovaleryl-CoA in the tricarboxylic acid cycle, leading to acetyl-CoA and thus the production of even-numbered FAs (Aepfler et al., 2019).
In natural AOM environments, there is circumstantial evidence for the presence of heterotrophic bacteria because of the abundance of branched FAs. Originally, different FA patterns have been described as originating from environments dominated by either ANME-1 or -2 but showing the presence of the same SRB partner (Blumenberg et al., 2004; Elvert et al., 2005; Niemann and Elvert, 2008). ANME-1 dominated AOM systems are related to lower methane flux and are dominated by aiC15:0 as well as other branched-chain FAs (Stadnitskaia et al., 2008), while ANME-2 dominated systems are indicated by the presence of monounsaturated C16:1ω5 and cyC17:0ω5,6 FAs at sites with high methane flux intensity (Elvert et al., 2003). Taking the results of our study into account, we suggest that the different FA patterns, particularly the larger amounts of branched and partly unsaturated fatty acids, originate from the activity of heterotrophic bacteria inhabiting the vicinity of AOM consortia. These heterotrophic bacteria effectively utilize available amino acids, such as valine, leucine, or isoleucine, which are derived from 13C-depleted proteins from AOM consortia necromass or AOM cell exudates/lysates, such as amino acids and acetate (Middelboe and Jørgensen, 2006; Takano et al., 2018; Yang et al., 2020; Hu et al., 2021). Thus, if amino acid-based carbon is available, especially under energy- and nutrient-limited conditions in ANME-1 dominated settings, bacterial heterotrophs will produce more branched FAs. Under natural conditions, such branched FAs become even more negative in δ13C values than FAs derived from the autotrophic SRB partner in AOM consortia (Elvert et al., 2005). Moreover, the activity of such heterotrophic bacteria may explain the difficulty of detecting AOM biomarkers in the sulfate methane transition zone (SMTZ) or their disappearance below the current SMTZ (Niemann et al., 2005; Biddle et al., 2006; Zhu et al., 2021) because AOM biomass is more labile and accessible to these degraders than recalcitrant background organic matter. As a result, they actively reshape different carbon pools and contribute to biogeochemical carbon cycling in anoxic marine sediments.
In addition, both AOM enrichment cultures contain archaea with potential heterotrophic metabolisms, including the members of the Bathyarchaeota, Thermoplasmatales, and Lokiarchaeota (Supplementary Table 3; Wegener et al., 2016; Krukenberg et al., 2018). All these three archaeal groups encode protein catabolism or have been cultured on proteinaceous substrates (Imachi et al., 2020; Yin et al., 2022). The metabolic activity of these microbes in the AOM37 culture is supported by the methane-independent incorporation of 13C-leu into IPL-derived Phy (Figure 5). Bathyarchaeota—formerly known as the Miscellaneous Crenarchaeotal Group (MCG)—are widespread in anoxic sediments. Based on their genomes, some Bathyarchaeota may be protein-degrading heterotrophs with acetyl-CoA centralized pathways for energy conservation (Lloyd et al., 2013). In the AOM50 culture, Bathyarchaeota, which accounts for approximately 10% of all cells (Supplementary Table 3), may be responsible for the trace incorporation of 13C into the CL-derived BP0 during the incubation (Figure 5). Another candidate for 13C-label incorporation from leucine is Thermoplasmatales, which occupy up to 4.9% of the total population in the Guaymas Basin cultures (Supplementary Table 3). The members of the Thermoplasmatales have been cultured with yeast extract as their carbon and energy sources (Itoh et al., 2007). They, therefore, may also be candidates for using leucine or its metabolized derivatives in the AOM enrichments. Lokiarchaeota, the third potential group, accounted for 0.4% of the whole population. Lokiarchaeota were only recently isolated and able to degrade amino acids via syntrophy, and they are likely to produce both archaeol and GDGTs as lipid membrane constituents (Imachi et al., 2020), which would be consistent with our study here. In summary, the low 13C labeling of archaeal lipids indicates that ancillary archaea play a small role in leucine turnover. However, given the widespread distribution of archaea and their postulated advantage over bacteria under conditions of severe energy stress, an archaeal contribution to the utilization of AOM-derived (dissolved) organic matter in the methane-laden sediments has to be taken into account (Biddle et al., 2006; Kubo et al., 2012; Yoshinaga et al., 2015).
Conclusion
Meso- and thermophilic AOM cultures from the Guaymas Basin were incubated with position-specifically labeled 13C-leu to investigate heterotrophic lipid formation by ancillary community members. Most of the 13C from leucine was incorporated into branched-chain and unsaturated FAs of heterotrophic bacteria, such as Anaerolineae or Spirochetes. No 13C-leu incorporation into FAs was observed for the cultured Ca. D. auxilii SRB representative, confirming that this partner bacterium is an autotroph. Combining our results with former environmental information of FA patterns of different AOM consortia indicates that bacterial heterotrophs thrive on 13C-depleted AOM necromass or cell exudates/lysates in the form of amino acids in the marine environment, addressing the frequently observed strong decline of AOM biomass and lipid biomarkers below current SMTZs. In addition, archaeol-based IPLs and some tetraether CLs showed minor methane-independent assimilation of 13C, suggesting that ancillary, potentially heterotrophic archaea, such as Bathyarchaeota, Thermoplasmatales, and Lokiarchaeota, are active. All these taxa are minor community members in our enrichment cultures but commonly appear in subsurface sediments and can thus be specialists for the recycling of necromass in anoxic hydrocarbon-rich habitats. The AOM cultures, with their limited microbial diversity, appear to be a promising source of materials for confirming the function of these mostly uncultured microorganisms through targeted cultivation.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
ME and Q-ZZ designed the research. Q-ZZ and GW performed the experiment. Q-ZZ analyzed the lipid data. Q-ZZ, ME, GW, and K-UH contributed to the discussion of the results and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the Deutsche Forschungsgemeinschaft (DFG) through the Gottfried Wilhelm Leibniz Prize awarded to K-UH (Hi 616-14-1) and DFG under Germany’s Excellence Initiative/Strategy through the Clusters of Excellence EXC 309 “The Ocean in the Earth System” (project no. 49926684) and EXC 2077 “The Ocean Floor – Earth’s Uncharted Interface” (project no. 390741601). Q-ZZ was sponsored by the China Scholarship Council (CSC) and Bremen International Graduate School for Marine Sciences (GLOMAR).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor SE declared a past co-authorship with one of the authors, GW.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Susanne Menger for assisting in the cultivation and Heidi Taubner for her help in measuring δ13C values of DIC. We are also thankful to Xavier Prieto and Jenny Wendt for their support in instrument maintenance. We also thank the editor SE and the reviewers SB and AS for their feedback on improving the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.912299/full#supplementary-material
References
Aepfler, R. F., Bühring, S. I., and Elvert, M. (2019). Substrate characteristic bacterial fatty acid production based on amino acid assimilation and transformation in marine sediments. FEMS Microbiol. Ecol. 95:fiz131. doi: 10.1093/femsec/fiz131
Biddle, J. F., Lipp, J. S., Lever, M. A., Lloyd, K. G., Sørensen, K. B., Anderson, R., et al. (2006). Heterotrophic archaea dominate sedimentary subsurface ecosystems off Peru. PNAS 103, 3846–3851. doi: 10.1073/pnas.0600035103
Blumenberg, M., Seifert, R., Reitner, J., Pape, T., and Michaelis, W. (2004). Membrane lipid patterns typify distinct anaerobic methanotrophic consortia. PNAS 101, 11111–11116. doi: 10.1073/pnas.0401188101
Boetius, A., Ravenschlag, K., Schubert, C. J., Rickert, D., Widdel, F., Gieseke, A., et al. (2000). A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407, 623–626. doi: 10.1038/35036572
Cord-Ruwisch, R. (1985). A quick method for the determination of dissolved and precipitated sulfides in cultures of sulfate-reducing bacteria. J. Microbiol. Methods 4, 33–36. doi: 10.1016/0167-7012(85)90005-3
Díaz-Pérez, A. L., Díaz-Pérez, C., and Campos-García, J. (2016). Bacterial l -leucine catabolism as a source of secondary metabolites. Rev. Environ. Sci. Biotechnol. 15, 1–29. doi: 10.1007/s11157-015-9385-3
Dombrowski, N., Teske, A. P., and Baker, B. J. (2018). Expansive microbial metabolic versatility and biodiversity in dynamic Guaymas Basin hydrothermal sediments. Nat. Commun. 9:4999. doi: 10.1038/s41467-018-07418-0
Dong, X., Greening, C., Brüls, T., Conrad, R., Guo, K., Blaskowski, S., et al. (2018). Fermentative Spirochaetes mediate necromass recycling in anoxic hydrocarbon-contaminated habitats. ISME J. 12, 2039–2050. doi: 10.1038/s41396-018-0148-3
Elvert, M., Boetius, A., Knittel, K., and Jørgensen, B. B. (2003). Characterization of specific membrane fatty acids as chemotaxonomic markers for sulfate-reducing bacteria involved in anaerobic oxidation of methane. Geomicrobiol. J. 20, 403–419. doi: 10.1080/01490450303894
Elvert, M., Hopmans, E. C., Treude, T., Boetius, A., and Suess, E. (2005). Spatial variations of methanotrophic consortia at cold methane seeps: implications from a high-resolution molecular and isotopic approach. Geobiology 3, 195–209. doi: 10.1111/j.1472-4669.2005.00051.x
Ganesan, B., Dobrowolsk, P., and Weimer, B. C. (2006). Identification of the leucine-to-2-methylbutyric acid catabolic pathway of Lactococcus lactis. Appl. Environ. Microbiol. 72, 4264–4273. doi: 10.1128/AEM.00448-06
Harwood, C. S., and Canale-Parola, E. (1981). Branched-chain amino acid fermentation by a marine spirochete: strategy for starvation survival. J. Bacteriol. 148, 109–116. doi: 10.1128/jb.148.1.109-116.1981
Henrichs, S. M., and Farrington, J. W. (1979). Amino acids in interstitial waters of marine sediments. Nature 279, 319–322. doi: 10.1038/279319a0
Heuer, V., Elvert, M., Tille, S., Krummen, M., Mollar, X. P., Hmelo, L. R., et al. (2006). Online δ13C analysis of volatile fatty acids in sediment/porewater systems by liquid chromatography-isotope ratio mass spectrometry. Limnol. Oceanogr-Meth. 4, 346–357.
Hinrichs, K.-U., Hayes, J. M., Sylva, S. P., Brewer, P. G., and DeLong, E. F. (1999). Methane-consuming archaebacteria in marine sediments. Nature 398, 802–805. doi: 10.1038/19751
Hinrichs, K.-U., Summons, R. E., Orphan, V., Sylva, S. P., and Hayes, J. M. (2000). Molecular and isotopic analysis of anaerobic methane-oxidizing communities in marine sediments. Org. Geochem. 31, 1685–1701. doi: 10.1016/S0146-6380(00)00106-6
Holler, T., Widdel, F., Knittel, K., Amann, R., Kellermann, M. Y., Hinrichs, K.-U., et al. (2011). Thermophilic anaerobic oxidation of methane by marine microbial consortia. ISME J. 5, 1946–1956. doi: 10.1038/ismej.2011.77
Hu, T., Luo, M., Xu, Y., Gong, S., and Chen, D. (2021). Production of labile protein-like dissolved organic carbon associated with anaerobic methane oxidization in the haima cold seeps, south china sea. Front. Microbiol. 8:1841. doi: 10.3389/fmars.2021.797084
Imachi, H., Nobu, M. K., Nakahara, N., Morono, Y., Ogawara, M., Takaki, Y., et al. (2020). Isolation of an archaeon at the prokaryote–eukaryote interface. Nature 577, 519–525. doi: 10.1038/s41586-019-1916-6
Itoh, T., Yoshikawa, N., and Takashina, T. (2007). Thermogymnomonas acidicola gen. nov., sp. nov., a novel thermoacidophilic, cell wall-less archaeon in the order thermoplasmatales, isolated from a solfataric soil in Hakone, Japan. Int. J. Syst. Evol. Microbiol. 57, 2557–2561. doi: 10.1099/ijs.0.65203-0
Jahn, U., Summons, R., Sturt, H., Grosjean, E., and Huber, H. (2004). Composition of the lipids of Nanoarchaeum Equitans and their origin from its host Ignicoccus sp. strain KIN4/I. Arch. Microbiol. 182, 404–413. doi: 10.1007/s00203-004-0725-x
Kaneda, T. (1977). Fatty acids of the genus Bacillus: an example of branched-chain preference. Bacteriol. Rev. 41, 391–418. doi: 10.1128/br.41.2.391-418.1977
Kellermann, M. Y., Wegener, G., Elvert, M., Yoshinaga, M. Y., Lin, Y.-S., Holler, T., et al. (2012). Autotrophy as a predominant mode of carbon fixation in anaerobic methane-oxidizing microbial communities. PNAS 109, 19321–19326. doi: 10.1073/pnas.1208795109
Kellermann, M. Y., Yoshinaga, M. Y., Wegener, G., Krukenberg, V., and Hinrichs, K.-U. (2016). Tracing the production and fate of individual archaeal intact polar lipids using stable isotope probing. Org. Geochem. 95, 13–20. doi: 10.1016/j.orggeochem.2016.02.004
Kirchman, D., K’nees, E., and Hodson, R. (1985). Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl. Environ. Microbiol. 49, 599–607. doi: 10.1128/aem.49.3.599-607.1985
Krukenberg, V., Harding, K., Richter, M., Glöckner, F. O., Gruber-Vodicka, H. R., Adam, B., et al. (2016). Candidatus desulfofervidus auxilii, a hydrogenotrophic sulfate-reducing bacterium involved in the thermophilic anaerobic oxidation of methane. Environ. Microbiol. 18, 3073–3091. doi: 10.1111/1462-2920.13283
Krukenberg, V., Riedel, D., Gruber-Vodicka, H. R., Buttigieg, P. L., Tegetmeyer, H. E., Boetius, A., et al. (2018). Gene expression and ultrastructure of meso- and thermophilic methanotrophic consortia. Environ. Microbiol. 20, 1651–1666. doi: 10.1111/1462-2920.14077
Kubo, K., Lloyd, K. G., Biddle, F. J., Amann, R., Teske, A., and Knittel, K. (2012). Archaea of the miscellaneous crenarchaeotal group are abundant, diverse and widespread in marine sediments. ISME J. 6, 1949–1965. doi: 10.1038/ismej.2012.37
Laso-Pérez, R., Krukenberg, V., Musat, F., and Wegener, G. (2018). Establishing anaerobic hydrocarbon-degrading enrichment cultures of microorganisms under strictly anoxic conditions. Nat. Protoc. 13, 1310–1330. doi: 10.1038/nprot.2018.030
Liang, B., Wang, L.-Y., Zhou, Z., Mbadinga, S. M., Zhou, L., Liu, J.-F., et al. (2016). High frequency of Thermodesulfovibrio spp. and Anaerolineaceae in association with Methanoculleus spp. in a long-term incubation of n-alkanes-degrading methanogenic enrichment culture. Front. Microbiol. 7:1431. doi: 10.3389/fmicb.2016.01431
Livermore, B. P., and Johnson, R. C. (1974). Lipids of the Spirochaetales: comparison of the lipids of several members of the genera Spirochaeta, Treponema, and Leptospira. J. Bacteriol. 120, 1268–1273.
Lloyd, K. G., Schreiber, L., Petersen, D. G., Kjeldsen, K. U., Lever, M. A., Steen, A. D., et al. (2013). Predominant archaea in marine sediments degrade detrital proteins. Nature 496, 215–218. doi: 10.1038/nature12033
Mårdén, P., Nyström, T., and Kjelleberg, S. (1987). Uptake of leucine by a marine Gram-negative heterotrophic bacterium during exposure to starvation conditions. FEMS Microbiol. Ecol. 3, 233–241. doi: 10.1111/j.1574-6968.1987.tb02361.x
McGlynn, S. E., Chadwick, G. L., Kempes, C. P., and Orphan, V. J. (2015). Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526, 531–535. doi: 10.1038/nature15512
Meador, T. B., Bowles, M., Lazar, C. S., Zhu, C., Teske, A., and Hinrichs, K.-U. (2015). The archaeal lipidome in estuarine sediment dominated by members of the miscellaneous crenarchaeotal group. Environ. Microbiol. 17, 2441–2458. doi: 10.1111/1462-2920.12716
Michaelis, W., Seifert, R., Nauhaus, K., Treude, T., Thiel, V., Blumenberg, M., et al. (2002). Microbial reefs in the Black Sea fueled by anaerobic oxidation of methane. Science 297, 1013–1015. doi: 10.1126/science.1072502
Middelboe, M., and Jørgensen, N. O. G. (2006). Viral lysis of bacteria: an important source of dissolved amino acids and cell wall compounds. J. Mar. Biol. Assoc. U. K. 86, 605–612. doi: 10.1017/S0025315406013518
Monticello, D. J., and Costilow, R. N. (1982). Interconversion of valine and leucine by Clostridium sporogenes. J. Bacteriol. 152, 946–949. doi: 10.1128/jb.152.2.946-949.1982
Niemann, H., and Elvert, M. (2008). Diagnostic lipid biomarker and stable carbon isotope signatures of microbial communities mediating the anaerobic oxidation of methane with sulphate. Org. Geochem. 39, 1668–1677. doi: 10.1016/j.orggeochem.2007.11.003
Niemann, H., Elvert, M., Hovland, M., Orcutt, B., Judd, A., Suck, I., et al. (2005). Methane emission and consumption at a North Sea gas seep (Tommeliten area). Biogeosciences 2, 335–351. doi: 10.5194/bg-2-335-2005
Niemann, H., Lösekann, T., de Beer, D., Elvert, M., Nadalig, T., Knittel, K., et al. (2006). Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 443, 854–858. doi: 10.1038/nature05227
Orphan, V. J., House, C. H., Hinrichs, K.-U., McKeegan, K. D., and DeLong, E. F. (2001). Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293, 484–487. doi: 10.1126/science.1061338
Orphan, V. J., House, C. H., Hinrichs, K.-U., McKeegan, K. D., and DeLong, E. F. (2002). Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. PNAS 99, 7663–7668. doi: 10.1073/pnas.072210299
Pancost, R. D., Hopmans, E. C., and Sinninghe Damsté, J. S. (2001). Archaeal lipids in Mediterranean cold seeps: molecular proxies for anaerobic methane oxidation. Geochim. Cosmochim. Acta 65, 1611–1627. doi: 10.1016/S0016-7037(00)00562-7
Paster, B. J. (2010). “Phylum XV. Spirochaetes Garrity and Holt 2001,” in Bergey’s Manual§of Systematic Bacteriology: Volume Four The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes, eds N. R. Krieg, J. T. Staley, D. R. Brown, B. P. Hedlund, B. J. Paster, N. L. Ward, et al. (New York, NY: Springer), 471–566. doi: 10.1007/978-0-387-68572-4_4
Pérez Castro, S., Borton, M. A., Regan, K., Hrabe de Angelis, I., Wrighton, K. C., Teske, A. P., et al. (2021). Degradation of biological macromolecules supports uncultured microbial populations in Guaymas Basin hydrothermal sediments. ISME J 15, 1–18. doi: 10.1038/s41396-021-01026-5
Preuß, A., Schauder, R., Fuchs, G., and Stichler, W. (1989). Carbon isotope fractionation by autotrophic bacteria with three different C02 fixation pathways. Z. Naturforsch. C 44, 397–402. doi: 10.1515/znc-1989-5-610
Reeburgh, W. S. (2007). Oceanic methane biogeochemistry. Chem. Rev. 107, 486–513. doi: 10.1021/cr050362v
Rosenkranz, F., Cabrol, L., Carballa, M., Donoso-Bravo, A., Cruz, L., Ruiz-Filippi, G., et al. (2013). Relationship between phenol degradation efficiency and microbial community structure in an anaerobic SBR. Water Res. 47, 6739–6749. doi: 10.1016/j.watres.2013.09.004
Rossel, P. E., Elvert, M., Ramette, A., Boetius, A., and Hinrichs, K.-U. (2011). Factors controlling the distribution of anaerobic methanotrophic communities in marine environments: evidence from intact polar membrane lipids. Geochim. Cosmochim. Acta 75, 164–184. doi: 10.1016/j.gca.2010.09.031
Rossel, P. E., Lipp, J. S., Fredricks, H. F., Arnds, J., Boetius, A., Elvert, M., et al. (2008). Intact polar lipids of anaerobic methanotrophic archaea and associated bacteria. Org. Geochem. 39, 992–999. doi: 10.1016/j.orggeochem.2008.02.021
Ruff, S. E., Biddle, J. F., Teske, A. P., Knittel, K., Boetius, A., and Ramette, A. (2015). Global dispersion and local diversification of the methane seep microbiome. PNAS 112, 4015–4020. doi: 10.1073/pnas.1421865112
Stadnitskaia, A., Ivanov, M. K., and Sinninghe Damsté, J. S. (2008). Application of lipid biomarkers to detect sources of organic matter in mud volcano deposits and post-eruptional methanotrophic processes in the Gulf of Cadiz. NE Atlantic. Mar. Geol. 255, 1–14. doi: 10.1016/j.margeo.2007.11.006
Sturt, H. F., Summons, R. E., Smith, K., Elvert, M., and Hinrichs, K.-U. (2004). Intact polar membrane lipids in prokaryotes and sediments deciphered by high-performance liquid chromatography/electrospray ionization multistage mass spectrometry—new biomarkers for biogeochemistry and microbial ecology. Rapid Commun. Mass Spectrom 18, 617–628. doi: 10.1002/rcm.1378
Suzuki, Y., Sasaki, T., Suzuki, M., Nogi, Y., Miwa, T., Takai, K., et al. (2005). Novel chemoautotrophic endosymbiosis between a member of the Epsilonproteobacteria and the hydrothermal-vent Gastropod Alviniconcha aff. hessleri (Gastropoda: Provannidae) from the Indian Ocean. Appl. Environ. Microbiol. 71, 5440–5450. doi: 10.1128/AEM.71.9.5440-5450.2005
Takano, Y., Chikaraishi, Y., Imachi, H., Miyairi, Y., Ogawa, N. O., Kaneko, M., et al. (2018). Insight into anaerobic methanotrophy from 13C/12C- amino acids and 14C/12C-ANME cells in seafloor microbial ecology. Sci. Rep. 8:14070. doi: 10.1038/s41598-018-31004-5
Thiel, V., Peckmann, J., Seifert, R., Wehrung, P., Reitner, J., and Michaelis, W. (1999). Highly isotopically depleted isoprenoids: molecular markers for ancient methane venting. Geochim. Cosmochim. Acta 63, 3959–3966. doi: 10.1016/S0016-7037(99)00177-5
Vishnuvardhan Reddy, S., Aspana, S., Tushar, D. L., and Sasikala, Ch, and Ramana, Ch (2013). Spirochaeta sphaeroplastigenens sp. nov., a halo-alkaliphilic, obligately anaerobic spirochaete isolated from soda lake Lonar. Int. J. Syst. Evol. Microbiol. 63, 2223–2228. doi: 10.1099/ijs.0.046292-0
Wegener, G., Gropp, J., Taubner, H., Halevy, I., and Elvert, M. (2021). Sulfate-dependent reversibility of intracellular reactions explains the opposing isotope effects in the anaerobic oxidation of methane. Sci. Adv. 7:eabe4939. doi: 10.1126/sciadv.abe4939
Wegener, G., Krukenberg, V., Riedel, D., Tegetmeyer, H. E., and Boetius, A. (2015). Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 526, 587–590. doi: 10.1038/nature15733
Wegener, G., Krukenberg, V., Ruff, S. E., Kellermann, M. Y., and Knittel, K. (2016). Metabolic capabilities of microorganisms involved in and associated with the anaerobic oxidation of methane. Front. Microbiol. 7:46. doi: 10.3389/fmicb.2016.00046
Wegener, G., Niemann, H., Elvert, M., Hinrichs, K.-U., and Boetius, A. (2008). Assimilation of methane and inorganic carbon by microbial communities mediating the anaerobic oxidation of methane. Environ. Microbiol. 10, 2287–2298. doi: 10.1111/j.1462-2920.2008.01653.x
Widdel, F., and Bak, F. (1992). “Gram-Negative Mesophilic Sulfate-Reducing Bacteria,” in The Prokaryotes: A Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Applications, eds A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (New York, NY: Springer), 3352–3378. doi: 10.1007/978-1-4757-2191-1_21
Wirsen, C. O., Sievert, S. M., Cavanaugh, C. M., Molyneaux, S. J., Ahmad, A., Taylor, L. T., et al. (2002). Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp. that produces filamentous sulfur. Appl. Environ. Microbiol. 68, 316–325. doi: 10.1128/AEM.68.1.316-325.2002
Yamada, T., Sekiguchi, Y., Hanada, S., Imachi, H., Ohashi, A., Harada, H., et al. (2006). Anaerolinea thermolimosa sp. nov., Levilinea saccharolytica gen. nov., sp. nov. and Leptolinea tardivitalis gen. nov., sp. nov., novel filamentous anaerobes, and description of the new classes Anaerolineae classis nov. and Caldilineae classis nov. in the bacterial phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 56, 1331–1340. doi: 10.1099/ijs.0.64169-0
Yamauchi, N. (2010). The pathway of leucine to mevalonate in halophilic archaea: efficient incorporation of leucine into isoprenoidal lipid with the involvement of isovaleryl-coa dehydrogenase in Halobacterium salinarum. Biosci. Biotechnol. Biochem. 74, 443–446. doi: 10.1271/bbb.90814
Yang, S., Lv, Y., Liu, X., Wang, Y., Fan, Q., Yang, Z., et al. (2020). Genomic and enzymatic evidence of acetogenesis by anaerobic methanotrophic archaea. Nat. Commun. 11:3941. doi: 10.1038/s41467-020-17860-8
Yin, X., Zhou, G., Cai, M., Zhu, Q.-Z., Richter-Heitmann, T., Aromokeye, D. A., et al. (2022). Catabolic protein degradation in marine sediments confined to distinct archaea. ISME J. 1–10. doi: 10.1038/s41396-022-01210-1
Yoshinaga, M. Y., Lazar, C. S., Elvert, M., Lin, Y.-S., Zhu, C., Heuer, V. B., et al. (2015). Possible roles of uncultured archaea in carbon cycling in methane-seep sediments. Geochim. Cosmochim. Acta 164, 35–52. doi: 10.1016/j.gca.2015.05.003
Keywords: anaerobic oxidation of methane, archaea, bacteria, heterotrophy, stable isotope probing, lipid biomarkers
Citation: Zhu Q-Z, Wegener G, Hinrichs K-U and Elvert M (2022) Activity of Ancillary Heterotrophic Community Members in Anaerobic Methane-Oxidizing Cultures. Front. Microbiol. 13:912299. doi: 10.3389/fmicb.2022.912299
Received: 04 April 2022; Accepted: 02 May 2022;
Published: 02 June 2022.
Edited by:
S. Emil Ruff, Marine Biological Laboratory (MBL), United StatesReviewed by:
Susma Bhattarai, IHE Delft Institute for Water Education, NetherlandsAxel Schippers, Federal Institute for Geosciences and Natural Resources, Germany
Copyright © 2022 Zhu, Wegener, Hinrichs and Elvert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing-Zeng Zhu, qzzhu@marum.de
 Qing-Zeng Zhu
Qing-Zeng Zhu Gunter Wegener
Gunter Wegener Kai-Uwe Hinrichs
Kai-Uwe Hinrichs Marcus Elvert
Marcus Elvert