Abstract
Cyclones have one of the greatest effects on the biodiversity of coral reefs and the associated species. But it is unknown how stochastic alterations in habitat structure influence metapopulation structure, connectivity and genetic diversity. From 1993 to 2018, the reefs of the Capricorn Bunker Reef group in the southern part of the Great Barrier Reef were impacted by three tropical cyclones including cyclone Hamish (2009, category 5). This resulted in substantial loss of live habitat-forming coral and coral reef fish communities. Within 6–8 years after cyclones had devastated, live hard corals recovered by 50–60%. We show the relationship between hard coral cover and the abundance of the neon damselfish (Pomacentrus coelestis), the first fish colonizing destroyed reefs. We present the first long-term (2008–2015 years corresponding to 16–24 generations of P. coelestis) population genetic study to understand the impact of cyclones on the meta-population structure, connectivity and genetic diversity of the neon damselfish. After the cyclone, we observed the largest change in the genetic structure at reef populations compared to other years. Simultaneously, allelic richness of genetic microsatellite markers dropped indicating a great loss of genetic diversity, which increased again in subsequent years. Over years, metapopulation dynamics were characterized by high connectivity among fish populations associated with the Capricorn Bunker reefs (2200 km2); however, despite high exchange, genetic patchiness was observed with annual strong genetic divergence between populations among reefs. Some broad similarities in the genetic structure in 2015 could be explained by dispersal from a source reef and the related expansion of local populations. This study has shown that alternating cyclone-driven changes and subsequent recovery phases of coral habitat can greatly influence patterns of reef fish connectivity. The frequency of disturbances determines abundance of fish and genetic diversity within species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cyclones are one of the most important key drivers of ecological heterogeneity and ecosystem function (Halford et al. 2004; Fabricius et al. 2008; Halford and Perret 2009; Roff et al. 2015; Puotinen et al. 2016). At the Great Barrier Reef, Australia, tropical storms and cyclones account for 48% of the respective estimated losses of coral cover followed by coral predation by crown of thorns starfish that caused 42% and coral bleaching 10% of coral loss (De’ath et al. 2012); the latter, however, has recently increased dramatically (Torda et al. 2018). Physical abrasion of coral cover, rolling and tossing of coral boulders and extensive movements of rubble all cause pronounced changes and destruction of a reef architecture. Storm-induced coral loss can have a devastating effect on coral species assemblages due to loss of shelter, habitat and food (Doherty et al. 1997; Woolsey et al. 2012). In contrast to these devastating effects, Connell (1978) predicted that intermediate disturbance such as storms might be beneficial to ecosystem functioning because disturbances create the highest diversity by maintaining coral reefs in a non-equilibrium state with newly appearing niches providing space and shelter to specialized species with high colonization potential. Soon after a severe disturbance, diversity is supposed to be low because the time for colonization is short; only those few species that can cope with reduced shelter, produce propagules fast and happen to be within dispersal range will colonize disturbed habitat after a cyclone.
While most ecologists have focused on changes in biodiversity and re-colonization potential after a disturbance event, the integration of evolutionary processes that have shaped species assemblages is much less considered. Yet, genetic variation within populations is the essence of evolution and may affect population viability and adaptability with consequences for the composition of associated communities, their metapopulation structure and ecosystem functioning (see for review Banks et al. 2013). In a modeling approach, Fraser et al. (2018) predicted that species with dispersal on small spatial scales, but rare on larger scales would show no spatial structure on small scales and strong structure on large scales. Furthermore, disturbance would influence the chance of genetic turnover in a population.
Our long-term study (2008–2015) on the coral reef fish Pomacentrus coelestis, a common species of the Great Barrier Reef, Australia, offers an ideal opportunity to test these predictions and to understand how disturbances can drive metapopulation dynamics and intraspecific structure. At the Capricorn Bunker Reef (CBR) group (Fig. 1) P. coelestis has a surprisingly short life cycle with an age-maxima of 127–160 days (Kingsford et al. 2017), which means a potential local population extirpation and a population turnover of two to three generations per year (corresponding to16–24 generations in 8 years from 2008–2015). P. coelestis prefer highly disturbed coral reefs (Doherty et al. 1996); the abundance of P. coelestis is generally low when percentage cover of live coral is high (Bell and Galzin 1984; Halford et al. 2004). After a cyclone event, abundance of most other fishes such as macro- and meso-predators of the families Serranidae and Lutjanidae that seek habitat complexity is greatly reduced; therefore, metapopulation structure and connectivity of P. coelestis is mostly driven by habitat availability (Jones et al. 2004).
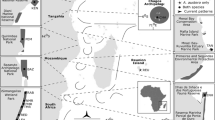
Map of reef sites in the Capricorn Bunker Reef Group combined with the population genetic structure of P. coelestis at 14 different reefs in 2015; from north to south: Broomfield, North West, Sykes, Heron, Wistari, Mast Head, One Tree, Lamont, Fitzroy, Llewellyn, Boult, Hoskyn, Fairfax, Lady Musgrave. Different colors of fish indicate significant genetic differences according to Dest values. Note, genetic differences between sampling sites at the same reef is indicated by two different colors
We aimed to understand how stochastic habitat appearance would influence metapopulation structure and genetic diversity. Our approach was to analyze the dynamics of the population genetic structure before and after a cyclone disturbance. We intended to reveal settlement and colonization at reefs affected by storm events and try to locate source-reef populations within the CBR group at the southern tip of the Great Barrier Reef, within an area of approx. 2200 km2.
Based on Banks et al. (2013) we tested following predictions after disturbances: (1) Losses of allelic diversity within populations were anticipated where mortality was high and population recovery came from a few in situ survivors or founding colonizers (particularly from a single source) (Beheregaray et al. 2003), and where ongoing immigration was low relative to the population growth rate (Pannell John and Charlesworth 2000). (2) there would be no loss of genetic diversity within disturbance-affected populations when survival was high (Suárez et al. 2012) or recovering populations recruited many individuals from multiple sources (Spear et al. 2012).
In our study, we specifically asked (1) How does the impact of a cyclone alter coral cover and abundance of P. coelestis? (2) Do stochastic disturbance events change genetic diversity in coral reef fish, and what is the time span of re-establishment of genetic diversity? (3) Do stochastic colonization events alter patterns of connectivity?
Materials and methods
Study area, assessing coral cover and cyclone damage
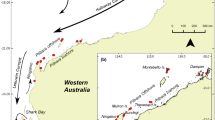
The Capricorn-Bunker group of reefs is located on the tropic of Capricorn about 100 km offshore in the southern part of the Great Barrier Reef, Australia (Fig. 1). Individual reefs are separated by 3–60 km. The southern Great Barrier Reef (GBR) is periodically impacted by cyclones and storms; in particular, a category 5 tropical cyclone (TC) Hamish hit the Capricorn Bunker reefs (CBR) in March 2009 (Online Resource Fig. S1). At One Tree Island (OTI) reef, benthic surveys of exposed sites documented a large reduction in live coral cover. Laminar corals in particular, such as Acropora hyacinthus, largely disappeared and filamentous algae cover increased by 30%, while lagoon and lee sites were not strongly affected (Woolsey et al. 2012). During the cyclone, OTI reef sustained southeasterly gale force winds (> 61.2 km h−1) for 18.5 h and swells > 6 m in height for 4 h. Two years after the cyclone, algal cover remained high and laminar corals had not recovered; probably, because of large wave action that was not directly related to cyclones in the area. From monitoring over 5 years, ocean mooring data revealed that OTI reef regularly experienced large swells (4–8 m). These swells from other storm events would have contributed to the physical disturbance of benthic biota and decline in coral cover recorded prior to and after TC Hamish (Woolsey et al. 2012).
Coral cover was recorded annually at One Tree Island from 2007 at a site on the North East slope (Site 1, 23°29.708 S, 152°05.887E). A replicate site 2, about 2 km from site 1, was monitored approximately biennially from 2010 (Site 2: 23°29.214 S, Long. 152°05.498E). Placement of the points was determined with 1–2 fin kicks and observed cover off the left hand corner of the slate. At each site, percentage cover was estimated along five transects (5 × 25 m), and point estimates were obtained for 20–35 points in each transect. This sample size gave a 95% chance of detecting any type of substratum that represented 10% or more of cover (Kingsford and Battershill 1998). P. coelestis are generally found in 6 to 20 m depth on the exposed sides of reefs. The five haphazardly placed transects, separated by meters, covered over 100 m of reef and provided an accurate estimate of P. coelestis in microhabitats that included live and dead coral.
Public domain data from the Australian Institute of Marine Science (AIMS) long-term monitoring provided benthic cover data at three sites on the exposed side (facing east) of Broomfield and Lady Musgrave reefs annually from 1993, OTI reef was surveyed annually 1993 to 2005, then biennially thereafter. Boult and Hoskyn reefs were surveyed biennially from 2004 onward; five transects were sampled at each site per reef (6–9 m deep). The reefs were all on the outer shelf of the Great Barrier Reef and form a N-S axis from Broomfield to Lady Musgrave (Fig. 1). Multi-reef comparisons of benthic cover were gained from 50 UW images from video frames (before 2005) or digital photographs (after 2005) and were taken at 1 m intervals along each transect. The percent cover of benthic organisms was estimated by identifying the organisms beneath five points projected onto each of 40 randomly selected images from 50 taken at each transect (3,000 points per reef, (Jonker et al. 2008)). Here, we present data on total hard coral cover, which include all coral morphologies, and tabulate acroporids, which are a major habitat forming corals, especially on the exposed sides of the CBR (Woolsey et al 2012). Abundance of the small pomacentrid P. coelestis was also counted in each of the fifteen 50 m transects (transect width was 1 m) per reef and year (i.e., data averaged for the 3 sites, n = 5). All transects were on the exposed slope at a depth of 6–9 m.
Fish collection
Almost annually from 2008 to 2015, we sampled P. coelestis from four to five reefs at the Capricorn Bunker reef group Fitzroy, Lamont, Heron, One Tree, Wistari. In 2015, we sampled at those and nine more reefs (Broomfield, North West, Sykes, Mast Head, Llewellyn, Boult, Hoskyn, Fairfax, Lady Musgrave) along the axis of the Capricorn Bunker Group covering a range of approx. 20 × 110 km (Fig. 1; Online Resource Table S1). We collected small P. coelestis by hand nets (95% were between 10 mm-35 mm SL) or with hand spears (5% of animals were > 35 mm) at 2–3 sites per reef separated by hundreds of meters to kilometers. This approach was used to decrease the probability of sampling related individuals and to gain a representative sample of the genetic diversity within each reef. Pectoral fins were clipped and stored in 99% ethanol until DNA extraction. Altogether we sampled 1992 P. coelestis, see Online Resource Table S1 for details. The methods were performed in accordance with relevant guidelines and regulations and approved by the Great Barrier Reef Marine Park Authority, Queensland Department of Primary Industry permits and the James Cook University Ethics Committee.
Genetic analyses
DNA was isolated from the samples using Chelex chelating resin (BioRad Chelex 100 Resin). We used up to seven previously described polymorphic DNA microsatellite markers: Pom3, Pom6, Pom21 and Pom25, AC37, LIST12, PA2.29 (Watts et al. 2004; Miller-Sims et al. 2005, 2008). PCR was carried out using approximately 100 ng of template DNA and the following cycle treatment; initial step of 5 min at 94ºC, followed by 35 cycles of 30 s at 94 ºC, 30 s at 44–46 ºC, and 1 min at 72 ºC, with a final extension step of 5 min at 72 ºC. Total reaction volume was 25 µL and contained 2.5 µL 10 X RED Taq Polymerase Buffer (Sigma, 10 mM Tris–HCl, pH 8.3, 50 mM KCl, 1.1 mM MgCl2 and 0.01% gelatin), 100 µM of each dNTP (Promega), 0.5 µM of both forward and reverse primer, and 0.5 U Taq polymerase (Sigma) 0.25 U RED Taq DNA Polymerase (Sigma). PCR fragments were separated and scored on a Beckman-Coulter CEQ™ 2000XL. For 2011 and 2012, we could not amplify the microsatellite PA229. When these years were part of an analysis we also excluded this marker for other populations. We compared the results with and without locus PA229. For genetic clustering, we used allele imputation on the raw data and compared the results when all individuals with missing alleles in all other loci were included (n = 1910) or excluded (n = 1685). This procedure allowed us to see if any results depended on missing data and allele imputation bias, rather than biological variance.
Data analysis
Expected and observed heterozygosity and FIS values were assessed using the program GENETIX (Belkhir et al. 1997) (Online Resource Table S4). Allelic richness was computed using FSTAT version 2.93 (Goudet 1995). Dest, but not FST values show the degree of genetic differentiation between populations based on multiallelic microsatellite marker (Jost 2008), Dest values ranging from 0 (no genetic differentiation) to 1 (no allele sharing: complete genetic differentiation were calculated by DEMEtics v. 0.8-7 (Gerlach et al. 2010).
We used the discriminant analysis of principal components (DAPC) (Jombart et al. 2010) implemented in the adegenet package (Jombart 2008) for the R software of the R Development Core Team (2009). DAPC is a multivariate method designed to identify and describe clusters of genetically similar individuals, providing assignment of individuals to groups, a visual assessment of between-population differentiation, and contribution of individual alleles to population structuring. The number of clusters was assessed using the R-based function find.clusters, which runs successive K-means clustering with increasing number of clusters (K) implemented in R. The chosen number of clusters was determined using the minimum number of clusters after which the Bayesian Information Criterion (BIC) increases or decreases by a negligible amount (see Online Resource Fig. S3). In order to see the effect of missing values on clustering, we used the recommended allele imputation method by Jombart et al. 2010 as implemented in DAPC (i.e., replacing NA alleles with mean allele frequencies), and we removed missing alleles from the data by using only individuals with complete information in the six loci (Pom3, Pom6, Pom21 and Pom25, AC37, LIST12).
All datasets generated and analyzed during this study are included in this published article (and in its supplementary information files). For comparison, we used also LEA: An R Package for Landscape and Ecological Association Studies for a structure like clustering algorithm (Frichot and François 2015). For imputation of missing data (NAs), we used the method ‘mode,’ which imputes NAs with the most likely genotype based on a first clustering approach; for comparison we also used ‘random’ which used allele frequencies to impute missing data similarly to DAPC; we also used this approach on the dataset with all missing allele information removed.
Results
Major disturbances of the Capricorn Bunker Reef group by Tropical Cyclone Hamish
The study area was impacted by a category 5 tropical cyclone (TC Hamish) hitting the CBR in March 2009, which caused widespread damage to coral habitats (Woolsey et al. 2012). Changes in the cover of live coral was substantial after the cyclone. At One Tree Island, the seascape on the exposed side of the reef changed from 42% to 18–25% total coral cover after the cyclone (Fig. 2a—cover at OTI reef; Fig. 3) and the effect size was even greater at other reefs in the Capricorn Bunker Group along the route of TC Hamish (Online Resource Fig. S1). Immediately after TC Hamish coral cover dropped at Broomfield, Boult, Hoskyn, One Tree and Lady Musgrave, and low live coral was sustained over several years (Fig. 2a, b, c; Table 1).
Relationship of coral cover and abundance of P. coelestis. Data are from the AIMS long-term monitoring program; data are mean values ± SE. a coral cover and abundance of P. coelestis (per 50 m2) at OTI One Tree Island reef from 2005 to 2019. Coral cover at a second site located ~ 2 km from NE Slope is presented as squares), b) coral cover and abundance of P. coelestis (per 50 m2) at Broomfield and c) Lady Musgrave reefs from 1992 to 2018; red arrows indicate when TC Hamish hit the reefs; Tropical storms Fran and Rewa hit the reefs in 1992 and 1993. Note that at Lady Musgrave reef P. coelestis population mostly increased already in 2010, which suggests that it might function as a source population
The average reduction in total hard coral from the 5 years before TC Hamish compared to the 5 years following was between 51 and 90% (Table1). The loss of tabulate coral was particularly clear, with intact tabulate acroporids (e.g., Acropora hyacinthus) all but disappearing from the exposed side of reefs with a 65–98% reduction in cover comparing the 5 years before TC Hamish with the 5 years after (Table 1). About 5 years after TC Hamish corals clearly recovered, but it took 8–10 years before coral cover reached the level prior to 2009. There was a series of cyclones that impacted the southern GBR and likely caused the damage to coral cover detected on seaward side of reefs in 1993 (cyclone Fran 1992, cyclone Rewa 1993; https://coast.noaa.gov/hurricanes/). A similar change from low to high hard coral cover was found from 1993 (when the survey started) to 2000 when coral cover was also observed (Fig. 2b).
The abundance of the short-lived fish P. coelestis increased rapidly at all reefs with a major decrease in coral cover after TC Hamish. When coral cover was high it corresponded with a major drop in abundance of P. coelestis (Fig. 2a, b, c; Table1); but there were always a few patches of suitable habitat where local groups of P. coelestis could prevail. In 2010, a year and five months after the cyclone, mean abundance of P. coelestis was highest at Lady Musgrave (107 per 50 m2, Fig. 2c) compared to more northern reefs Hoskyn (41 per 50 m2), Boult (34 per 50 m2) and Broomfield (51 per 50 m2, Fig. 2b).
Temporal and spatial dynamics in the genetic structure
The genetic structure of P. coelestis showed large differences between adjacent reef populations and between different years. In detail, Figs. 1 and 4a show—for better understanding simplified—color-coded P. coelestis reef populations based on statistically significant Dest values; different colors indicate statistically significant different populations. Detailed pairwise comparisons (Dest values) and statistics are presented in Online Resource Tables S2 and S3.
Genetic structure of P. coelestis reef populations from 2008 to 2015. a Different colors of fish symbols indicate significant genetic differences of pairwise Dest values; details are shown in Table S2 and S3, b) Pairwise genetic comparisons (Dest, median) among four reef populations (Fitzroy, Heron, Lamont and OTI); * Wilcoxon signed rank test, z2008–2010 = -−1.99, df = 5, p = 0.046; z2008-2015 = 1.78, df = 5, p= 0.093; ns nonsignificant; indicating that after the cyclone genetic differences between reef populations increased. c Mean allelic richness of four reef populations (Fitzroy, Heron, Lamont and OTI) considering six different SSR markers; different letters indicate statistical significance. Note: We use the term ‘clade’ to address differences among reef populations based on Dest values; but we use the term ‘cluster’ indicating differences within and between reef populations based on DAPC analyses (see below).
The biggest genetic differences between populations were observed before and after the cyclone hit the reefs (between 2008 and 2010) with Dest values ranging from 0.14 (Heron2008 and Heron2010) to 0.29 (Lamont2008 and Lamont2010). Figure 4b shows yearly differences based on mean Dest values among four reef populations, which were almost consistently sampled from 2008 to 2015 (Fitzroy, Heron, Lamont and OTI); values of 2014 were excluded when fish were only sampled at Heron and Fitzroy; in 2009 no fish were sampled; detailed Dest values see Online Resource Table S3.
We analyzed the temporal and spatial dynamics of the genetic structure with a Discriminant Analysis of Principle Components (DAPC), as implemented in the R package adegenet (Jombart 2008; Jombart et al. 2010) shown in Online Resource Fig. S2 and Fig. S3. The Bayesian information criterion (BIC) was used to choose a most parsimonious model for grouping analysis. The lowest BIC in the distribution of BIC over K clusters was found for 14 clusters. We finally used a minimum number of eight different clusters K, as increasing the numbers of K beyond K = 8 decreases the BIC only marginally (Online Resource Fig. S3a). All reef populations from 2008 to 2015 were visualized in the DAPC chart (Online Resource Fig. S3b), a flowchart showing the relative number of individuals belonging to each cluster K per population (Fig. 5) and a bar plot showing the probability of each individual belonging to one of the eight specified clusters (Online Resource Fig. S2). Overall, the analysis showed that the P. coelestis populations were not defined by reef, but each reef population consisted of a mix of up to eight genotypic clusters. Additionally, a DAPC flowchart (Fig. 5) validated for different years showed that two major changes occurred: the first change occurred between 2008 and 2010 coinciding with TC Hamish; here, especially clusters 3, 5, 6 and 7, which were frequent at all reefs in 2008, diminished and cluster 1 and 8 dominated reef populations in 2010 until 2012, which probably corresponds to the dominance of the ‘yellow’ clade in Fig. 4. The next substantial change occurred after 2012. In view of Fig. 5, the genetic cluster composition in this third period after 2012 looks similar to the first in 2008, with the frequency of clusters 3, 4, 5, and 7 increasing again while cluster 1 and 8 decreased.
Flowchart of a DAPC analysis designed to identify and describe clusters of genetically related individuals per population. The chosen number of clusters (K = 8) was determined using the minimum number of clusters after which the BIC increases or decreases by a negligible amount, see Online Resource Fig. S3. Red and blue lines show major changes of cluster composition, red line corresponds to TC Hamish, blue line corresponds to increasing recovery of coral
In 2015, when we sampled 14 reef populations in the CBR group, we re-found the ‘purple’ clade that we detected in 2008 only at OTI reef (‘purple’ in Fig. 4, which might correspond to an absence of clusters 1 and 8 in OTI2008 shown in Fig. 5). In 2015, this clade dominated most of the southern Capricorn Bunker reefs from Lady Musgrave (probably the source reef, see above) to Llewellyn and the northern reefs Broomfield, North West Island. Two additional clades were found at Wistari, Sykes and Mast Head Island Reef, the ‘turquoise’ clade that differed from the ‘purple’ clade by a maximum Dest value = 0.33. Interestingly, at OTI reef in 2015 a further clade ‘green’ showed up (Fig. 1, 4) differing from all other reefs 2015 by a Dest = 0.17.
To summarize the main results, we observed rapid changes between years especially large ones after the cyclone and likely spill-over effects of specific clades, e.g., Fig. 1 wide distribution of the ‘purple’ clade from Lady Musgrave to most other reefs of the GBR and as a further example the ‘yellow’ clade from Heron 2008 to the OTI, Lamont, Fitzroy and Wistari reefs in the following years (Fig. 4a), which can all easily be explained by increased stock size and connectivity based on larval dispersal.
We tested the data also with different approaches in DAPC using the method ‘ward’ instead of ‘Kmeans’ for the clustering approach prior to DAPC and used both the AIC instead of BIC for comparison. To test for the influence of missing data and different clustering approaches we run DAPC analyses. They showed similar results indicating that the population structure is not based on missing data (NAs). We also removed all NA data and got again similar results. We added information for number of clusters K = 2–7 (see Online Resource Figs. S4a–g). LEA with different statistics and imputation methods did not show the same results as the DAPC. In most cases, K = 1 was the most likely number of clusters, closely followed by K = 3. But the bar chart of membership probability when using K > 1 was very similar to the results of the DAPC, with allele imputation methods on the full dataset and with missing allele individuals and loci removed from the study. Therefore, we considered K > 1 as reliable.
Dynamics of genetic diversity
Following Schmeller et al. (2018) who proposed a suite of essential biodiversity variables (EBVs), we focused on allelic richness through time as a measure of genetic diversity that is indicative of a population’s long-term potential for adaptability and persistence. As an indicator for genetic diversity, we determined the dynamics of allelic richness for genetic markers in all years and four reef populations of Fitzroy, Heron, Lamont and OTI reef. We found that allelic richness significantly differed among years (Kruskall Wallis H = 15.31, df = 5, p = 0.009) (Fig. 4c), being the highest in 2008 before the storm and dropped in following years 2010 and 2011. Based on Benjamini and Hochberg corrections (1995) for multiple comparisons, only allelic richness in 2011 was significantly different from 2008 (z = 3.184, df = 5, p = 0.002).
We compared number of alleles (per locus divided by number of sampled individuals to account for different sample size) between two different size classes from 4 reefs from 2013 and found that small individuals (10–25 mm SL) had significantly fewer alleles than medium-sized individuals (25–35 mm SL) (Wilcoxon Signed Rank Test; Z = 28, n = 27, p = 0.0009).
Discussion
Impact of TC Hamish on reef fish population
We provide strong evidence that the increased availability of suitable habitat caused by TC Hamish greatly influenced patterns of abundance of P. coelestis, the size of potential source populations and pathways of connectivity. P. coelestis is a species that responds quickly to changes in habitat type (sensu ‘habitat responder’ Kingsford et al. 2019) and is most abundant on dead coral pavement and rubble, a habitat that abounds after major disturbances (see also Bell and Galzin 1984; Doherty et al. 1996; Halford et al. 2004). Once coral cover increases, an assemblage of other coral reef fishes colonize this habitat. The decline in abundance of P. coelestis could be explained by them being displaced by other fish rather than actually avoiding live corals (Matis et al. 2018). Tropical cyclones and rough seas that originate from greater latitudes, impacted many square kilometers of reef, particularly on the windward side (Halford et al. 2004; Woolsey et al. 2012). Before TC Hamish devastated the Capricorn Bunker Reef group in March 2009, hard corals covered 70–80% of the reefs at OTI reef, making it an unsuitable habitat for P. coelestis (Fig. 2a). Correspondingly, the number of counted P. coelestis was at a minimum of two animals per 50 m2 (Fig. 2a). Similar situations with high coral cover and low numbers of P. coelestis were observed on other reefs, e.g., Broomfield and Lady Musgrave before the cyclone. After the cyclone, reefs in the impact zone had their coral cover reduced by 51–98% (Table 1). The populations of P. coelestis responded with an immediate increase in abundance of up to 120 per 50 m2 at Lady Musgrave reef and at OTI reef after a delay to more than 160 per 50 m2 (Table 1, Fig. 2a, c). Densities of P. coelestis at Lady Musgrave reef increased quicker when compared to, e.g., OTI reefs and Bromfield reefs (Fig. 2a–c), which we suggest made Lady Musgrave reef a potential source reef of the widespread ‘purple’ clade. Replenishment can be rapid due to a long spawning season, short generation time and high survival on arrival due to a loss of live coral habitat and associated predators. Replenishment can occur through two primary mechanisms. (1) It may be assisted by input from other source reefs (as we argue post TC Hamish). Further, (2) from a local low population base, with rapid spawning a high proportion of fish returning to their natal reef or settle at nearby reefs (Wolanski and Kingsford 2014). The coral cover at effected reefs recovered fast; we have already observed a significant increase in some types of hard corals 7 years after TC Hamish hit the reefs (Fig. 3). When corals covered the reef again by almost 60%, P. coelestis abundance decreased quickly at all reefs (Table 1).
Connectivity and metapopulation structure before and after TC Hamish
The temporal and spatial analyses of the population genetic structure of P. coelestis provide insights into the effects of a major storm event on connectivity, metapopulation structure and genetic diversity. In 2008, the genetic structure in our core reef group populations (Fitzroy, Heron and Lamont) were similar, only the OTI reef population differed from the other three reef populations by Dest values: 0.05 to 0.1. The wide distribution of the ‘turquoise’ clade shows connectivity among reef populations at Fitzroy, Heron and Lamont that are 10 to 20 km apart. However, locally at OTI a genetically different clade ‘purple’ was found indicating that despite connectivity by larval dispersal, genotypes were not homogenously mixed among reefs. However, the DAPC analysis clearly shows that the color codes are over-simplifying the situation. A clade did not consist of a single genotypic cluster but of a mix of clusters, which differed in frequency.
After TC Hamish in 2010, the population genetic structure had changed significantly in all four reef populations. New clusters (1 and 8) became predominant, while the others 3 and 5 decreased or even disappeared until 2013. From 2013 on a significant genetic change took place; clusters and their frequency were more similar to the pre-cyclone situation. Also the DAPC analysis (Fig. 5 and Fig. 2S) indicated this similarity: again as in 2008, clusters 1 and 8 decreased and clusters 3, 4, 5, and 6 increased in frequency.
We provided strong ecological evidence that the P. coelestis population at Lady Musgrave showed the fastest recovery in less than 1 year (2010) after TC Hamish. Here, the abundance of P. coelestis increased from a trace to a mean of 113 per 50 m2 within 17 months. This large population would have had the potential for strong natal recruitment and subsequent significant spill over (Russ and Alcala 1996) in a northerly direction serving as a source population for other reefs that were 80 km or more to the north, e.g., reefs OTI, Fitzroy, Lamont and Heron. However, it took some years to dominate these reef-based populations; the first purple clade was detected in 2014 at this group of reefs 3–4 years after the perturbation caused by TC Hamish.
The most interesting result is that a sudden habitat increase, and therefore a higher settlement probability did not result in genetic homogeneity of recruits at all reefs. Shoals of larval fish randomly searching for settlement habitat at the end of their dispersal phase and finding it everywhere should lead to a more random distribution. Indeed, P. coelestis settled earlier after the cyclone than before the cyclone (Planktonic Larval Duration: year 2011: 20.4 days ± 0.39 SE, n = 127, year 2008: 23.3 days ± 0.36 SE, n = 159, chi2 = 34.0, p < 0.0001; data unpublished), which may suggest that a pre-settlement ‘search phase’ (sensu: Sih and Kingsford 2016) may be shorter when it is easier to find an appropriate settlement habitat after the cyclone. If settlement habitats were rarer, juveniles would need more time to search and would travel longer distances before they find a settlement place.
However, this quicker settlement did not lead to a random and homogeneous genotype distribution, but to genetic patchiness, which has also been observed in other fishes (Pujolar et al. 2006; Selkoe et al. 2006; Selwyn et al. 2016). This idea proposes that a large variance in reproductive success, causes surviving offspring to exhibit some characteristics of bottle-necked populations, namely larvae come from a small fraction of the adult population (Selkoe et al. 2010). Such patchiness in the population structure is certainly influenced by the short life cycle of P. coelestis. If larvae from a different genotypic cluster succeed, a completely new genetically different population will take over fast because the previous reef population has already died off. Kin structured dispersal and orientation behavior (Gerlach et al. 2007; Mouritsen et al. 2013; Paris et al. 2013; Bottesch et al. 2016) would support this pattern. But even in longer-lived coral reef fishes, high dynamics in the genetic structure and genetic differences among reef fish populations have been observed on a small geographic scale. Doherty et al. (1995) found that within the Great Barrier Reef three species of damselfish: Chromis atripectoralis, and Stegastes nigricans showed genetic differences on the 10 km scale of similar magnitude to differences found at the 1000 km scale. Temporal genetic differentiation has also been shown on the 10 km scale in the damselfish species Dascyllus trimaculatus (Bernardi et al. 2001) and in the same region in the cardinalfish Ostorhinchus doederleini (Gerlach et al. 2007, 2016). We explain genetic divergence observed among reef populations with range expansions via serial founder effects, but not by genetic drift or selection. Genetic differences should be interpreted as stochastic genetic patchiness caused by successful recruits from a limited number of parents that managed to produce their offspring at the right time and place. This resulted in a rapid population expansion of a small group of founders that may consist of kin-structured dispersing larvae (Ottmann et al. 2016). This might explain why we do not observe a genetically homogeneous population, see Horne’s (2014) review on barrier effects on the genetic divergence in the Indo-Pacific coral reef fauna.
Despite a large increase in suitable habitat caused by TC Hamish, allelic richness declined in all reef populations in 2010 (Fig. 4c) especially in highly polymorphic markers. The observed loss of allelic richness suggests that after a storm these newly emerging habitats were not colonized by multiple sources of animals from adjacent reefs, but by few founders with reduced genetic diversity and reduced number of alleles relative to the parental generation. This loss of alleles might later be countered by gene flow induced by migrants arriving from the source population carrying different alleles, and therefore, gene flow is a force that may recover allelic richness (Lacy 1987). Firstly, the cyclone had killed many fish and destroyed shelter, but had soon enlarged the cover of suitable habitats (i.e., dead coral and rubble), leading to a rapid population expansion of P. coelestis with a time-delayed increase in allelic richness.
Outlook for the future
Ocean warming directly threatens coral reefs through heat stress, but also boosting the energy of cyclones causing major coral destructions. According to the State of the Climate Report 2018 (http://www.csiro.au), fewer tropical cyclones are projected for the Australian region, on average. However, an increased proportion of intense cyclones is expected. As a habitat responder, P. coelestis will temporarily benefit from new and empty habitat. However, as soon as the corals are recovering, the mortality rate of P. coelestis will increase along with the availability of shelter for predators. Our results on P. coelestis could be generalized to the entire coral reef fish community, which will colonize the reef following a reduction in P. coelestis numbers and experience a similar situation, i.e., sudden habitat availability that favors successful founder groups of settlers. Based on our results, we predict that selection will increase to favor species that mature fast so that they can reproduce multiple times before their habitat is destroyed again. Species with longer maturation time and with special habitat requirements will find it increasingly difficult to maintain a population or colonize reefs successfully if habitat structure alters substantially over increasingly shorter periods.
References
Banks SC, Cary GJ, Smith AL, Davies ID, Driscoll DA, Gill AM, Lindenmayer DB, Peakall R (2013) How does ecological disturbance influence genetic diversity? Trends Ecol Evol 28:670–679
Beheregaray LB, Ciofi C, Geist D, Gibbs JP, Caccone A, Powell JR (2003) Genes Record a Prehistoric Volcano Eruption in the Galápagos. Science 302:75–75
Belkhir K, Borsa P, Goudet J, Chikhi L, Bonhomme F (1997) Genetix v. 3.0, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome et Populations, CNRS UPR 9060, Université Montpellier 2, Montpellier
Bell JD, Galzin R (1984) Influence of live coral cover on coral-reef fish communities. Mar Ecol Prog Ser 15:265–274
Benjamini Y, Hochberg Y (1995) Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Statist Soc B 57:289–300
Bernardi G, Holbrook SJ, Schmitt RJ (2001) Gene flow at three spatial scales in a coral reef fish, the three-spot dascyllus, Dascyllus trimaculatus. Mar Biol 138:457–465
Bottesch M, Gerlach G, Halbach M, Bally A, Kingsford MJ, Mouritsen H (2016) A magnetic compass that might help coral reef fish larvae return to their natal reef. Curr Biol 26:R1266–R1267
Connell JH (1978) Diversity in Tropical Rain Forests and Coral Reefs. Science 199:1302–1310
De’ath G, Fabricius KE, Sweatman H, Puotinen M (2012) The 27-year decline of coral cover on the Great Barrier Reef and its causes. Proc Nat Acad Sci USA 109:17995–17999
Doherty PJ, Planes S, Mather P (1995) Gene Flow and Larval Duration in 7 Species of Fish from the Great-Barrier-Reef. Ecology 76:2373–2391
Doherty P, Kingsford M, Booth D, Carleton J (1996) Habitat selection before settlement by Pomacentrus coelestis. Mar Freshwater Res 47:391–399
Doherty PJ, Meekan M, Miller IR, Osborne K, Thompson AA (1997) Catastrophic loss of coral cover from reefs in the southern Great Barrier Reef and the impact on fish recruitment. Proc Nat Acad Sci USA 1:1005–1010
Fabricius KE, De’ath G, Puotinen ML, Done T, Cooper TF, Burgess SC (2008) Disturbance gradients on inshore and offshore coral reefs caused by a severe tropical cyclone. Limnol Oceanogr 53:690–704
Fraser CI, Davies ID, Bryant D, Waters JM (2018) How disturbance and dispersal influence intraspecific structure. J Ecol 106:1298–1306
Frichot E, François O (2015) LEA: An R package for landscape and ecological association studies. Methods Ecol Evol 6:925–929
Gerlach G, Atema J, Kingsford MJ, Black KP, Miller-Sims V (2007) Smelling home can prevent dispersal of reef fish larvae. Proc Natl Acad Sci USA 104:858–863
Gerlach G, Jueterbock A, Kraemer P, Deppermann J, Harmand P (2010) Calculations of population differentiation based on G(ST) and D: forget G(ST) but not all of statistics! Mol Ecol 19:3845–3852
Gerlach G, Atema J, Raupach MJ, Deister F, Müller A, Kingsford MJ (2016) Cryptic species of cardinalfish with evidence for old and new divergence. Coral Reefs 35:437–450
Goudet J (1995) Fstat version 1.2: a computer program to calculate Fstatistics. J Heredity 86:485–486
Halford AR, Perret J (2009) Patterns of recovery in catastrophically disturbed reef fish assemblages. Mar Ecol Prog Ser 383:261–272
Halford A, Cheal AJ, Ryan D, Williams DM (2004) Resilience to large-scale disturbance in coral and fish assemblages on the Great Barrier Reef. Ecology 85:1892–1905
Horne JB (2014) Thinking outside the barrier: neutral and adaptive divergence in Indo-Pacific coral reef faunas. Evol Ecol 28:991–1002
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94
Jones GP, McCormick MI, Srinivasan M, Eagle JV (2004) Coral decline threatens fish biodiversity in marine reserves. Proc Natl Acad Sci USA 101:8251–8253
Jonker M, Johns K, Osborne K (2008) Surveys of benthic reef communities using underwater digital photography and counts of juvenile corals. In: Science AIoM (ed) Long-term Monitoring of the Great Barrier Reef, Townsville
Kingsford MJ, Battershill CN (1998) Studying Temperate Marine Environments: a handbook for ecologists. University of Canterbury Press, Christchurch, New Zealand
Kingsford MJ, O’Callaghan MD, Liggins L, Gerlach G (2017) The short-lived neon damsel Pomacentrus coelestis: implications for population dynamics. J Fish Biol 90:2041–2059
Kingsford MJ, Syms C, Srinivasan M, Jones GP (2019). In: Hutchings P, Kingsford M, Hoegh Guldberg O (eds) Great Barrier Reef: Biology, Environment and Management. CSIRO Publishing and CRC Press
Lacy RC (1987) Loss of Genetic Diversity from Managed Populations: Interacting Effects of Drift, Mutation, Immigration, Selection, and Population Subdivision. Cons Biol 1:143–158
Matis PA, Donelson JM, Bush S, Fox RJ, Booth DJ (2018) Temperature influences habitat preference of coral reef fishes: Will generalists become more specialised in a warming ocean? Glob Change Biol 24:3158–3169
Miller-Sims V, Atema J, Kingsford MJ, Gerlach G (2005) DNA microsatellite primers in the neon damselfish (Pomacentrus coelestis). Mol Ecol 5:424–426
Miller-Sims VC, Gerlach G, Kingsford MJ, Atema J (2008) Dispersal in the spiny damselfish, Acanthochromis polyacanthus, a coral reef fish species without a larval pelagic stage. Mol Ecol 17:5036–5048
Mouritsen H, Atema J, Kingsford MJ, Gerlach G (2013) Sun Compass Orientation Helps Coral Reef Fish Larvae Return to Their Natal Reef. PLoS ONE 8:e66039
Ottmann D, Grorud-Colvert K, Sard NM, Huntington BE, Banks MA, Sponaugle S (2016) Long-term aggregation of larval fish siblings during dispersal along an open coast. Proc Nat Acad Sci USA 113:14067–14072
Pannell John R, Charlesworth B (2000) Effects of metapopulation processes on measures of genetic diversity. Phil Trans R Soc Lond Ser B 355:1851–1864
Paris CB, Atema J, Irisson J-O, Kingsford M, Gerlach G, Guigand CM (2013) Reef Odor: A Wake Up Call for Navigation in Reef Fish Larvae. PLoS ONE 8:e72808
Pujolar JM, Maes GE, Volckaert FAM (2006) Genetic patchiness among recruits in the European eel Anguilla anguilla. Mar Ecol Prog Ser 307:209–217
Puotinen M, Maynard JA, Beeden R, Radford B, Williams GJ (2016) A robust operational model for predicting where tropical cyclone waves damage coral reefs. Sci Rep 6:26009
Roff G, Doropoulos C, Zupan M, Rogers A, Steneck RS, Golbuu Y, Mumby PJ (2015) Phase shift facilitation following cyclone disturbance on coral reefs. Oecologia 178:1193–1203
Russ GR, Alcala AC (1996) Do marine reserves export adult fish biomass? Evidence from Apo Island, central Phillipines. Mar Ecol Prog Ser 132:1–9
Schmeller DS, Weatherdon LV, Loyau A, Bondeau A, Brotons L, Brummitt N, Geijzendorffer IR, Haase P, Kuemmerlen M, Martin CS, Mihoub JB, Rocchini D, Saarenmaa H, Stoll S, Regan EC (2018) A suite of essential biodiversity variables for detecting critical biodiversity change. Biol Rev 93:55–71
Selkoe KA, Gaines SD, Caselle JE, Warner RR (2006) Current shifts and kin aggregation explain genetic patchiness in fish recruits. Ecology 87:3082–3094
Selkoe KA, Watson JR, White C, Ben Horin T, Iacchei M, Mitarai S, Siegel DA, Gaines SD, Toonen RJ (2010) Taking the chaos out of genetic patchiness: seascape genetics reveals ecological and oceanographic drivers of genetic patterns in three temperate reef species. Mol Ecol 19:3708–3726
Selwyn JD, Hogan JD, Downey-Wall AM, Gurski LM, Portnoy DS, Heath DD (2016) Kin-aggregations explain chaotic genetic patchiness, a commonly observed genetic pattern in a marine fish. PLoS ONE 11:e0153381
Sih TL, Kingsford MJ (2016) Near-reef elemental signals in the otoliths of settling Pomacentrus amboinensis (Pomacentridae). Coral Reefs 35:303–315
Spear SF, Crisafulli CM, Storfer A (2012) Genetic structure among coastal tailed frog populations at Mount St. Helens is moderated by post-disturbance management. Ecol Appl 22:856–869
Suárez NM, Betancor E, Fregel R, Rodríguez F, Pestano J (2012) Genetic signature of a severe forest fire on the endangered Gran Canaria blue chaffinch (Fringilla teydea polatzeki). Conserv Genet 13:499–507
Team RDC (2009) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria
Torda G, Sambrook K, Cross P, Sato Y, Bourne DG, Lukoschek V, Hill T, Torras Jorda G, Moya A, Willis BL (2018) Decadal erosion of coral assemblages by multiple disturbances in the Palm Islands, central Great Barrier Reef. Sci Rep 8:11885–11885
Watts PC, Veltsos P, Soffa BJ, Gill AB, Kemp SJ (2004) Polymorphic microsatellite loci in the black and gold chromis Neoglyphidodon nigroris (Teleostei: Pomacentridae). Mol Ecol 5:93–95
Wolanski E, Kingsford MJ (2014) Oceanographic and behavioural assumptions in models of the fate of coral and coral reef fish larvae. J R Soc Interface 11:2014.0209
Woolsey E, Bainbridge SJ, Kingsford MJ, Byrne M (2012) Impacts of cyclone Hamish at One Tree Reef: integrating environmental and benthic habitat data. Mar Biol 159:793–803
Acknowledgements
We thank The Australian Institute of Marine Science (AIMS) Long-Term Monitoring Program (LTMP) for providing data on coral cover and abundance of P. coelestis and Hugh Sweatman and Mike Emslie for additional comments on the manuscript. We thank Mark O’Callaghan and Andreas Bally for assistance with sample collection; Susanne Wallenstein and Anke Mueller for helping to process samples and Andreas Bally providing figures. This work was funded by the German Science Foundation (Ge 842/6-1, FOR 2716 and SFB 1372) to GG and funding from the ARC Centre of Excellence for Coral Reef Studies to MJK.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Topic editor Morgan S. Pratchett
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gerlach, G., Kraemer, P., Weist, P. et al. Impact of cyclones on hard coral and metapopulation structure, connectivity and genetic diversity of coral reef fish. Coral Reefs 40, 999–1011 (2021). https://doi.org/10.1007/s00338-021-02096-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00338-021-02096-9